Single-molecule and super-resolution imaging of transcription in living bacteria
- PMID: 28414097
- PMCID: PMC5670121
- DOI: 10.1016/j.ymeth.2017.04.001
Single-molecule and super-resolution imaging of transcription in living bacteria
Abstract
In vivo single-molecule and super-resolution techniques are transforming our ability to study transcription as it takes place in its native environment in living cells. This review will detail the methods for imaging single molecules in cells, and the data-analysis tools which can be used to extract quantitative information on the spatial organization, mobility, and kinetics of the transcription machinery from these experiments. Furthermore, we will highlight studies which have applied these techniques to shed new light on bacterial transcription.
Keywords: Live cells; PALM; Quantitative microscopy; Single-particle tracking; Super-resolution imaging; Transcription.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

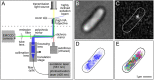
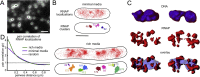
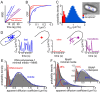

References
Publication types
MeSH terms
Substances
Grants and funding
- BB/N018656/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/J00054X/1 /BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 261227/ERC_/European Research Council/International
- BB/H01795X/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 204684/Z/16/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources

