Structure of the PRC2 complex and application to drug discovery
- PMID: 28414199
- PMCID: PMC5519257
- DOI: 10.1038/aps.2017.7
Structure of the PRC2 complex and application to drug discovery
Abstract
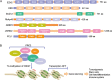
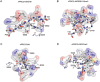
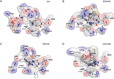
The polycomb repressive complexes 2 (PRC2) complex catalyzes tri-methylation of histone H3 lysine 27 (H3K27), a repressive chromatin marker associated with gene silencing. Overexpression and mutations of PRC2 are found in a wide variety of cancers, making the catalytic activity of PRC2 an important target of cancer therapy. This review highlights recent structural breakthroughs of the human PRC2 complex bound to the H3K27 peptide and a small molecule inhibitor, which provide critically needed insight into PRC2-targeted drug discovery.
Figures



Similar articles
-
Small Molecule Approaches for Targeting the Polycomb Repressive Complex 2 (PRC2) in Cancer.J Med Chem. 2020 Dec 24;63(24):15344-15370. doi: 10.1021/acs.jmedchem.0c01344. Epub 2020 Dec 7. J Med Chem. 2020. PMID: 33283516 Review.
-
Polycomb repressive complex 2 structure with inhibitor reveals a mechanism of activation and drug resistance.Nat Commun. 2016 Apr 28;7:11384. doi: 10.1038/ncomms11384. Nat Commun. 2016. PMID: 27122193 Free PMC article.
-
Structure, mechanism, and regulation of polycomb-repressive complex 2.J Biol Chem. 2018 Sep 7;293(36):13805-13814. doi: 10.1074/jbc.R117.800367. Epub 2017 Sep 14. J Biol Chem. 2018. PMID: 28912274 Free PMC article. Review.
-
An allosteric PRC2 inhibitor targeting the H3K27me3 binding pocket of EED.Nat Chem Biol. 2017 Apr;13(4):381-388. doi: 10.1038/nchembio.2304. Epub 2017 Jan 30. Nat Chem Biol. 2017. PMID: 28135235
-
Embryonic Ectoderm Development (EED) as a Novel Target for Cancer Treatment.Curr Top Med Chem. 2021;21(31):2771-2777. doi: 10.2174/1568026621666210920154942. Curr Top Med Chem. 2021. PMID: 34544341 Review.
Cited by
-
Telomere dysfunction cooperates with epigenetic alterations to impair murine embryonic stem cell fate commitment.Elife. 2020 Apr 16;9:e47333. doi: 10.7554/eLife.47333. Elife. 2020. PMID: 32297856 Free PMC article.
-
Hypermethylation of CDKN2A CpG island drives resistance to PRC2 inhibitors in SWI/SNF loss-of-function tumors.Cell Death Dis. 2024 Nov 5;15(11):794. doi: 10.1038/s41419-024-07109-3. Cell Death Dis. 2024. PMID: 39500892 Free PMC article.
-
C10ORF12 modulates PRC2 histone methyltransferase activity and H3K27me3 levels.Acta Pharmacol Sin. 2019 Nov;40(11):1457-1465. doi: 10.1038/s41401-019-0247-3. Epub 2019 Jun 11. Acta Pharmacol Sin. 2019. PMID: 31186533 Free PMC article.
-
Inhibition of EED-mediated histone methylation alleviates neuroinflammation by suppressing WNT-mediated dendritic cell migration.J Neuroinflammation. 2025 Apr 1;22(1):97. doi: 10.1186/s12974-025-03429-z. J Neuroinflammation. 2025. PMID: 40169990 Free PMC article.
-
Unravelling the impact of the chromobox proteins in human cancers.Cell Death Dis. 2025 Apr 2;16(1):238. doi: 10.1038/s41419-025-07585-1. Cell Death Dis. 2025. PMID: 40175347 Free PMC article. Review.
References
-
- Copeland RA, Olhava EJ, Scott MP. Targeting epigenetic enzymes for drug discovery. Curr Opin Chem Biol 2010; 14: 505–10. - PubMed
-
- Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov 2009; 8: 724–32. - PubMed
-
- Bauge C, Bazille C, Girard N, Lhuissier E, Boumediene K. Histone methylases as novel drug targets: developing inhibitors of EZH2. Future Med Chem 2014; 6: 1943–65. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

