Inherited GINS1 deficiency underlies growth retardation along with neutropenia and NK cell deficiency
- PMID: 28414293
- PMCID: PMC5409070
- DOI: 10.1172/JCI90727
Inherited GINS1 deficiency underlies growth retardation along with neutropenia and NK cell deficiency
Abstract
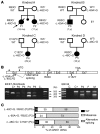
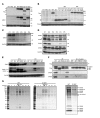
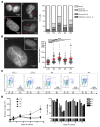
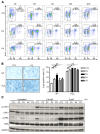
Inborn errors of DNA repair or replication underlie a variety of clinical phenotypes. We studied 5 patients from 4 kindreds, all of whom displayed intrauterine growth retardation, chronic neutropenia, and NK cell deficiency. Four of the 5 patients also had postnatal growth retardation. The association of neutropenia and NK cell deficiency, which is unusual among primary immunodeficiencies and bone marrow failures, was due to a blockade in the bone marrow and was mildly symptomatic. We discovered compound heterozygous rare mutations in Go-Ichi-Ni-San (GINS) complex subunit 1 (GINS1, also known as PSF1) in the 5 patients. The GINS complex is essential for eukaryotic DNA replication, and homozygous null mutations of GINS component-encoding genes are embryonic lethal in mice. The patients' fibroblasts displayed impaired GINS complex assembly, basal replication stress, impaired checkpoint signaling, defective cell cycle control, and genomic instability, which was rescued by WT GINS1. The residual levels of GINS1 activity reached 3% to 16% in patients' cells, depending on their GINS1 genotype, and correlated with the severity of growth retardation and the in vitro cellular phenotype. The levels of GINS1 activity did not influence the immunological phenotype, which was uniform. Autosomal recessive, partial GINS1 deficiency impairs DNA replication and underlies intra-uterine (and postnatal) growth retardation, chronic neutropenia, and NK cell deficiency.
Conflict of interest statement
Figures


Comment in
- Defects in DNA replication hit NK cells and neutrophils
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

