Transforming activity and therapeutic targeting of C-terminal-binding protein 2 in Apc-mutated neoplasia
- PMID: 28414304
- PMCID: PMC5561459
- DOI: 10.1038/onc.2017.106
Transforming activity and therapeutic targeting of C-terminal-binding protein 2 in Apc-mutated neoplasia
Abstract
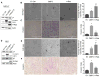
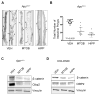
Overexpression of the transcriptional coregulators C-terminal binding proteins 1 and 2 (CtBP1 and 2) occurs in many human solid tumors and is associated with poor prognosis. CtBP modulates oncogenic gene expression programs and is an emerging drug target, but its oncogenic role is unclear. Consistent with this oncogenic potential, exogenous CtBP2 transformed primary mouse and human cells to anchorage independence similarly to mutant H-Ras. To investigate CtBP's contribution to in vivo tumorigenesis, Apcmin/+ mice, which succumb to massive intestinal polyposis, were bred to Ctbp2+/- mice. CtBP interacts with adenomatous polyposis coli (APC) protein, and is stabilized in both APC-mutated human colon cancers and Apcmin/+ intestinal polyps. Ctbp2 heterozygosity increased the median survival of Apcmin/+ mice from 21 to 48 weeks, and reduced polyp formation by 90%, with Ctbp2+/- polyps exhibiting reduced levels of β-catenin and its oncogenic transcriptional target, cyclin D1. CtBP's potential as a therapeutic target was studied by treating Apcmin/+ mice with the CtBP small-molecule inhibitors 4-methylthio-2-oxobutyric acid and 2-hydroxy-imino phenylpyruvic acid, both of which reduced polyposis by more than half compared with vehicle treatment. Phenocopying Ctbp2 deletion, both Ctbp inhibitors caused substantial decreases in the protein level of Ctbp2, as well its oncogenic partner β-catenin, and the effects of the inhibitors on CtBP and β-catenin levels could be modeled in an APC-mutated human colon cancer cell line. CtBP2 is thus a druggable transforming oncoprotein critical for the evolution of neoplasia driven by Apc mutation.
Conflict of interest statement
The authors have declared that no conflict of interest exists.
Figures

References
-
- Turner J, Crossley M. The CtBP family: enigmatic and enzymatic transcriptional co-repressors. Bio Essays News Rev Mol Cell Dev Biol. 2001;23:683–690. - PubMed
-
- Zhang C, Gao C, Xu Y, Zhang Z. CtBP2 could promote prostate cancer cell proliferation through c-Myc signaling. Gene. 2014;546:73–79. - PubMed
-
- Barroilhet L, Yang J, Hasselblatt K, Paranal RM, Ng S-K, Rauh-Hain JA, et al. C-terminal binding protein-2 regulates response of epithelial ovarian cancer cells to histone deacetylase inhibitors. Oncogene. 2013;32:3896–3903. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

