Initial sites of hepadnavirus integration into host genome in human hepatocytes and in the woodchuck model of hepatitis B-associated hepatocellular carcinoma
- PMID: 28414318
- PMCID: PMC5520499
- DOI: 10.1038/oncsis.2017.22
Initial sites of hepadnavirus integration into host genome in human hepatocytes and in the woodchuck model of hepatitis B-associated hepatocellular carcinoma
Abstract
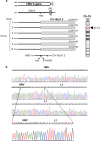
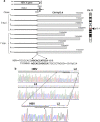
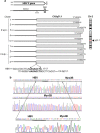
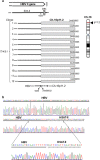
Hepatitis B virus (HBV) and the closely related woodchuck hepatitis virus (WHV) are potent carcinogens that trigger development of primary hepatocellular carcinoma (HCC). The initial sites of hepadnavirus-host genome integration, their diversity and kinetics of formation can be central to virus persistence and the initiation and progression of HCC. To recognize the nature of the very early virus-host interactions, we explored de novo infection of human hepatocyte-like HepaRG cells with authentic HBV and naive woodchucks with WHV. HepaRG were analyzed from several minutes post exposure to HBV onwards, whereas woodchuck liver biopsies at 1 or 3 h and 6 weeks post infection with WHV. Inverse PCR and clonal sequencing of the amplicons were applied to identify virus-host genomic junctions. HBV and WHV DNA and their replication intermediates became detectable in one hour after virus exposure. Concomitantly, HBV DNA integration into various host genes was detected. Notably, junctions of HBV X gene with retrotransposon sequences, such as LINE1 and LINE2, became prominent shortly after infection. In woodchucks, insertion of WHV X and preS sequences into host genome was evident at 1 and 3 h post infection (h.p.i.), confirming that hepadnavirus under natural conditions integrates into hepatocyte DNA soon after invasion. The HBV and WHV X gene enhancer II/core promotor sequence most often formed initial junctions with host DNA. Moreover, multiple virus-virus DNA fusions appeared from 1 h.p.i. onwards in both infected hepatocytes and woodchuck livers. In summary, HBV DNA integrates almost immediately after infection with a variety of host's sequences, among which tandemly repeating non-coding DNAs are common. This study revealed that HBV can engage mobile genetic elements from the beginning of infection to induce pro-oncogenic perturbations throughout the host genome. Such swift virus insertion was also evident in natural hepadnaviral infection in woodchucks.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Kinetics of DNA damage repair response accompanying initial hepadnavirus-host genomic integration in woodchuck hepatitis virus infection of hepatocyte.Cancer Genet. 2020 Jun;244:1-10. doi: 10.1016/j.cancergen.2020.02.001. Epub 2020 Feb 5. Cancer Genet. 2020. PMID: 32062411
-
Replication of naturally occurring woodchuck hepatitis virus deletion mutants in primary hepatocyte cultures and after transmission to naive woodchucks.J Virol. 2001 Apr;75(8):3811-8. doi: 10.1128/JVI.75.8.3811-3818.2001. J Virol. 2001. PMID: 11264370 Free PMC article.
-
Superinfection with woodchuck hepatitis virus strain WHVNY of livers chronically infected with strain WHV7.J Virol. 2015 Jan;89(1):384-405. doi: 10.1128/JVI.02361-14. Epub 2014 Oct 15. J Virol. 2015. PMID: 25320318 Free PMC article.
-
Application of the woodchuck animal model for the treatment of hepatitis B virus-induced liver cancer.World J Gastrointest Oncol. 2021 Jun 15;13(6):509-535. doi: 10.4251/wjgo.v13.i6.509. World J Gastrointest Oncol. 2021. PMID: 34163570 Free PMC article. Review.
-
The Initial Hepatitis B Virus-Hepatocyte Genomic Integrations and Their Role in Hepatocellular Oncogenesis.Int J Mol Sci. 2023 Oct 3;24(19):14849. doi: 10.3390/ijms241914849. Int J Mol Sci. 2023. PMID: 37834296 Free PMC article. Review.
Cited by
-
Hepatitis B Virus DNA Integration, Chronic Infections and Hepatocellular Carcinoma.Microorganisms. 2021 Aug 23;9(8):1787. doi: 10.3390/microorganisms9081787. Microorganisms. 2021. PMID: 34442866 Free PMC article. Review.
-
Systematically Exploring the Antitumor Mechanisms of Core Chinese Herbs on Hepatocellular Carcinoma: A Computational Study.Evid Based Complement Alternat Med. 2020 Sep 15;2020:2396569. doi: 10.1155/2020/2396569. eCollection 2020. Evid Based Complement Alternat Med. 2020. PMID: 33014099 Free PMC article.
-
Identification of antibodies cross-reactive with woodchuck immune cells and activation of virus-specific and global cytotoxic T cell responses by anti-PD-1 and anti-PD-L1 in experimental chronic hepatitis B and persistent occult hepadnaviral infection.Front Microbiol. 2022 Dec 6;13:1011070. doi: 10.3389/fmicb.2022.1011070. eCollection 2022. Front Microbiol. 2022. PMID: 36560951 Free PMC article.
-
Deciphering the Origin and Evolution of Hepatitis B Viruses by Means of a Family of Non-enveloped Fish Viruses.Cell Host Microbe. 2017 Sep 13;22(3):387-399.e6. doi: 10.1016/j.chom.2017.07.019. Epub 2017 Aug 31. Cell Host Microbe. 2017. PMID: 28867387 Free PMC article.
-
Impact of Hepatitis B Virus Genetic Variation, Integration, and Lymphotropism in Antiviral Treatment and Oncogenesis.Microorganisms. 2020 Sep 24;8(10):1470. doi: 10.3390/microorganisms8101470. Microorganisms. 2020. PMID: 32987867 Free PMC article. Review.
References
-
- World Health Organization. Hepatitis B fact sheet. Revised July. World Health Organization: Geneva, Switzerland, 2015, Available http://www.who.int/mediacentre/factsheets/fs204/en/. Accessed: 10 April 2016.
-
- Wollersheim M, Debelka U, Hofschneider PH. A transactivating function encoded in the hepatitis B virus X gene is conserved in the integrated state. Oncogene 1988; 3: 545–552. - PubMed
-
- Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol 2016; 64: S84–S101. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

