Sequence variability of the respiratory syncytial virus (RSV) fusion gene among contemporary and historical genotypes of RSV/A and RSV/B
- PMID: 28414749
- PMCID: PMC5393888
- DOI: 10.1371/journal.pone.0175792
Sequence variability of the respiratory syncytial virus (RSV) fusion gene among contemporary and historical genotypes of RSV/A and RSV/B
Erratum in
-
Correction: Sequence variability of the respiratory syncytial virus (RSV) fusion gene among contemporary and historical genotypes of RSV/A and RSV/B.PLoS One. 2017 Jun 28;12(6):e0180623. doi: 10.1371/journal.pone.0180623. eCollection 2017. PLoS One. 2017. PMID: 28658276 Free PMC article.
Abstract
Background: The fusion (F) protein of RSV is the major vaccine target. This protein undergoes a conformational change from pre-fusion to post-fusion. Both conformations share antigenic sites II and IV. Pre-fusion F has unique antigenic sites p27, ø, α2α3β3β4, and MPE8; whereas, post-fusion F has unique antigenic site I. Our objective was to determine the antigenic variability for RSV/A and RSV/B isolates from contemporary and historical genotypes compared to a historical RSV/A strain.
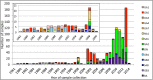
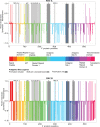
Methods: The F sequences of isolates from GenBank, Houston, and Chile (N = 1,090) were used for this analysis. Sequences were compared pair-wise to a reference sequence, a historical RSV/A Long strain. Variability (calculated as %) was defined as changes at each amino acid (aa) position when compared to the reference sequence. Only aa at antigenic sites with variability ≥5% were reported.
Results: A total of 1,090 sequences (822 RSV/A and 268 RSV/B) were analyzed. When compared to the reference F, those domains with the greatest number of non-synonymous changes included the signal peptide, p27, heptad repeat domain 2, antigenic site ø, and the transmembrane domain. RSV/A subgroup had 7 aa changes in the antigenic sites: site I (N = 1), II (N = 1), p27 (N = 4), α2α3β3β4(AM14) (N = 1), ranging in frequency from 7-91%. In comparison, RSV/B had 19 aa changes in antigenic sites: I (N = 3), II (N = 1), p27 (N = 9), ø (N = 4), α2α3β3β4(AM14) (N = 1), and MPE8 (N = 1), ranging in frequency from 79-100%.
Discussion: Although antigenic sites of RSV F are generally well conserved, differences are observed when comparing the two subgroups to the reference RSV/A Long strain. Further, these discrepancies are accented in the antigenic sites in pre-fusion F of RSV/B isolates, often occurring with a frequency of 100%. This could be of importance if a monovalent F protein from the historical GA1 genotype of RSV/A is used for vaccine development.
Conflict of interest statement
Figures

References
-
- Peret TC, Hall CB, Hammond GW, Piedra PA, Storch GA, Sullender WM, et al. Circulation patterns of group A and B human respiratory syncytial virus genotypes in 5 communities in North America. J Infect Dis. 2000;181: 1891–6. doi: 10.1086/315508 - DOI - PubMed
-
- Anderson LJ, Bingham P, Hierholzer JC. Neutralization of respiratory syncytial virus by individual and mixtures of F and G protein monoclonal antibodies. J Virol. 1988;62: 4232–8. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=253856&tool=pm... - PMC - PubMed
-
- Tan L, Coenjaerts FEJ, Houspie L, Viveen MC, van Bleek GM, Wiertz EJHJ, et al. The comparative genomics of human respiratory syncytial virus subgroups A and B: genetic variability and molecular evolutionary dynamics. J Virol. American Society for Microbiology (ASM); 2013;87: 8213–26. doi: 10.1128/JVI.03278-12 - DOI - PMC - PubMed
-
- PATH. RSV Vaccine Snapshot [Internet]. 2016 [cited 10 Jan 2017]. Available: http://www.path.org/vaccineresources/details.php?i=1562
-
- Johnson PR, Spriggs MK, Olmsted RA, Collins PL. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc Natl Acad Sci U S A. 1987;84: 5625–9. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=298915&tool=pm... - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

