Transcriptional networks are associated with resistance to Mycobacterium tuberculosis infection
- PMID: 28414762
- PMCID: PMC5393882
- DOI: 10.1371/journal.pone.0175844
Transcriptional networks are associated with resistance to Mycobacterium tuberculosis infection
Abstract
Rationale: Understanding mechanisms of resistance to M. tuberculosis (M.tb) infection in humans could identify novel therapeutic strategies as it has for other infectious diseases, such as HIV.
Objectives: To compare the early transcriptional response of M.tb-infected monocytes between Ugandan household contacts of tuberculosis patients who demonstrate clinical resistance to M.tb infection (cases) and matched controls with latent tuberculosis infection.
Methods: Cases (n = 10) and controls (n = 18) were selected from a long-term household contact study in which cases did not convert their tuberculin skin test (TST) or develop tuberculosis over two years of follow up. We obtained genome-wide transcriptional profiles of M.tb-infected peripheral blood monocytes and used Gene Set Enrichment Analysis and interaction networks to identify cellular processes associated with resistance to clinical M.tb infection.
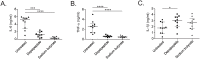
Measurements and main results: We discovered gene sets associated with histone deacetylases that were differentially expressed when comparing resistant and susceptible subjects. We used small molecule inhibitors to demonstrate that histone deacetylase function is important for the pro-inflammatory response to in-vitro M.tb infection in human monocytes.
Conclusions: Monocytes from individuals who appear to resist clinical M.tb infection differentially activate pathways controlled by histone deacetylase in response to in-vitro M.tb infection when compared to those who are susceptible and develop latent tuberculosis. These data identify a potential cellular mechanism underlying the clinical phenomenon of resistance to M.tb infection despite known exposure to an infectious contact.
Conflict of interest statement
Figures



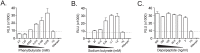
References
-
- WHO (2015) Global Tuberculosis Report.
-
- Flynn JL, Goldstein MM, Chan J, Triebold KJ, Pfeffer K, et al. (1995) Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity 2: 561–572. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

