The Epstein-Barr virus miR-BHRF1-1 targets RNF4 during productive infection to promote the accumulation of SUMO conjugates and the release of infectious virus
- PMID: 28414785
- PMCID: PMC5413087
- DOI: 10.1371/journal.ppat.1006338
The Epstein-Barr virus miR-BHRF1-1 targets RNF4 during productive infection to promote the accumulation of SUMO conjugates and the release of infectious virus
Abstract
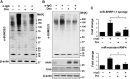
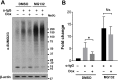
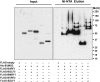
Post-translational modification by the Small Ubiquitin-like Modifier (SUMO) regulates a variety of cellular functions, and is hijacked by viruses to remodel the host cell during latent and productive infection. Here we have monitored the activity of the SUMO conjugation machinery in cells productively infected with Epstein-Barr virus (EBV). We found that SUMO2/3 conjugates accumulate during the late phase of the productive virus cycle, and identified several viral proteins as bone fide SUMOylation substrates. Analysis of the mechanism involved in the accumulation of SUMOylated proteins revealed upregulation of several components of the SUMO-conjugation machinery and post-transcriptional downregulation of the SUMO-targeted ubiquitin ligase RNF4. The latter effect was mediated by selective inhibition of RNF4 protein expression by the viral miR-BHRF1-1. Reconstitution of RNF4 in cells expressing an inducible miR-BHRF1-1 sponge or a miR-BHRF1-1 resistant RNF4 was associated with reduced levels of early and late viral proteins and impaired virus release. These findings illustrate a novel strategy for viral interference with the SUMO pathway, and identify the EBV miR-BHRF1-1 and the cellular RNF4 as regulators of the productive virus cycle.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








Similar articles
-
E1B-55K-Mediated Regulation of RNF4 SUMO-Targeted Ubiquitin Ligase Promotes Human Adenovirus Gene Expression.J Virol. 2018 Jun 13;92(13):e00164-18. doi: 10.1128/JVI.00164-18. Print 2018 Jul 1. J Virol. 2018. PMID: 29695423 Free PMC article.
-
Role of RNF4 in the ubiquitination of Rta of Epstein-Barr virus.J Biol Chem. 2013 May 3;288(18):12866-79. doi: 10.1074/jbc.M112.413393. Epub 2013 Mar 15. J Biol Chem. 2013. PMID: 23504328 Free PMC article.
-
Epstein-Barr virus encoded microRNAs target SUMO-regulated cellular functions.FEBS J. 2014 Nov;281(21):4935-50. doi: 10.1111/febs.13040. Epub 2014 Sep 30. FEBS J. 2014. PMID: 25205475
-
Genetics of Epstein-Barr virus microRNAs.Semin Cancer Biol. 2014 Jun;26:52-9. doi: 10.1016/j.semcancer.2014.02.002. Epub 2014 Mar 3. Semin Cancer Biol. 2014. PMID: 24602823 Review.
-
Manipulation of ubiquitin/SUMO pathways in human herpesviruses infection.Rev Med Virol. 2016 Nov;26(6):435-445. doi: 10.1002/rmv.1900. Epub 2016 Aug 22. Rev Med Virol. 2016. PMID: 27550835 Review.
Cited by
-
Bioinformatics analysis of miRNA and mRNA expression profiles to reveal the key miRNAs and genes in osteoarthritis.J Orthop Surg Res. 2021 Jan 19;16(1):63. doi: 10.1186/s13018-021-02201-2. J Orthop Surg Res. 2021. PMID: 33468167 Free PMC article.
-
The Epstein-Barr virus deubiquitinating enzyme BPLF1 regulates the activity of topoisomerase II during productive infection.PLoS Pathog. 2021 Sep 20;17(9):e1009954. doi: 10.1371/journal.ppat.1009954. eCollection 2021 Sep. PLoS Pathog. 2021. PMID: 34543352 Free PMC article.
-
Changes in SUMO-modified proteins in Epstein-Barr virus infection identifies reciprocal regulation of TRIM24/28/33 complexes and the lytic switch BZLF1.PLoS Pathog. 2023 Jul 6;19(7):e1011477. doi: 10.1371/journal.ppat.1011477. eCollection 2023 Jul. PLoS Pathog. 2023. PMID: 37410772 Free PMC article.
-
Human immunodeficiency virus type 1 impairs sumoylation.Life Sci Alliance. 2022 Feb 18;5(6):e202101103. doi: 10.26508/lsa.202101103. Print 2022 Jun. Life Sci Alliance. 2022. PMID: 35181598 Free PMC article.
-
A genome-wide screen of Epstein-Barr virus proteins that modulate host SUMOylation identifies a SUMO E3 ligase conserved in herpesviruses.PLoS Pathog. 2018 Jul 6;14(7):e1007176. doi: 10.1371/journal.ppat.1007176. eCollection 2018 Jul. PLoS Pathog. 2018. PMID: 29979787 Free PMC article.
References
-
- Desterro JM, Rodriguez MS, Kemp GD, Hay RT. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J Biol Chem. 1999;274(15):10618–24. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

