Speciation in progress? A phylogeographic study among populations of Hemitrichia serpula (Myxomycetes)
- PMID: 28414791
- PMCID: PMC5393559
- DOI: 10.1371/journal.pone.0174825
Speciation in progress? A phylogeographic study among populations of Hemitrichia serpula (Myxomycetes)
Abstract
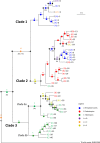
Myxomycetes (plasmodial slime molds, Amoebozoa) are often perceived as widely distributed, confounding to the "everything is everywhere" hypothesis. To test if gene flow within these spore-dispersed protists is restricted by geographical barriers, we chose the widespread but morphologically unmistakable species Hemitrichia serpula for a phylogeographic study. Partial sequences from nuclear ribosomal RNA genes (SSU) revealed 40 ribotypes among 135 specimens, belonging to three major clades. Each clade is dominated by specimens from a certain region and by one of two morphological varieties which can be differentiated by SEM micrographs. Partial sequences of the protein elongation factor 1 alpha (EF1A) showed each clade to possess a unique combination of SSU and EF1A genotypes. This pattern is best explained assuming the existence of several putative biospecies dominating in a particular geographical region. However, occasional mismatches between molecular data and morphological characters, but as well heterogeneous SSU and heterozygous EF1A sequences, point to ongoing speciation. Environmental niche models suggest that the putative biospecies are rather restricted by geographical barriers than by macroecological conditions. Like other protists, myxomycetes seem to follow the moderate endemicity hypothesis and are in active speciation, which is most likely shaped by limited gene flow and reproductive isolation.
Conflict of interest statement
Figures





References
-
- Skaloud P, Rindi F. (2013) Ecological differentiation of cryptic species within an asexual protist morphospecies: a case study of filamentous green alga Klebsormidium (Streptophyta). J Eukaryot Microbiol 60: 350–362. doi: 10.1111/jeu.12040 - DOI - PubMed
-
- Degerlund M, Huseby S, Zingone A, Sarno D, Landfald B. (2012) Functional diversity in cryptic species of Chaetoceros socialis Lauder (Bacillariophyta). J Plankton Res 34: 416–431.
-
- Halling RE, Sdmundson TDW, Neves MA. (2008) Pacific boletes: Implications for biogeographic relationships. Mycol Res 112: 437–447. doi: 10.1016/j.mycres.2007.11.021 - DOI - PubMed
-
- Lara E, Belbhari L. (2011) SSU rRNA reveals major trends in oomycete evolution. Fungal Divers 49: 93–100.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

