Semantic prioritization of novel causative genomic variants
- PMID: 28414800
- PMCID: PMC5411092
- DOI: 10.1371/journal.pcbi.1005500
Semantic prioritization of novel causative genomic variants
Abstract
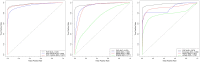
Discriminating the causative disease variant(s) for individuals with inherited or de novo mutations presents one of the main challenges faced by the clinical genetics community today. Computational approaches for variant prioritization include machine learning methods utilizing a large number of features, including molecular information, interaction networks, or phenotypes. Here, we demonstrate the PhenomeNET Variant Predictor (PVP) system that exploits semantic technologies and automated reasoning over genotype-phenotype relations to filter and prioritize variants in whole exome and whole genome sequencing datasets. We demonstrate the performance of PVP in identifying causative variants on a large number of synthetic whole exome and whole genome sequences, covering a wide range of diseases and syndromes. In a retrospective study, we further illustrate the application of PVP for the interpretation of whole exome sequencing data in patients suffering from congenital hypothyroidism. We find that PVP accurately identifies causative variants in whole exome and whole genome sequencing datasets and provides a powerful resource for the discovery of causal variants.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

