DNA barcoding evaluation and implications for phylogenetic relationships in Lauraceae from China
- PMID: 28414813
- PMCID: PMC5393608
- DOI: 10.1371/journal.pone.0175788
DNA barcoding evaluation and implications for phylogenetic relationships in Lauraceae from China
Abstract
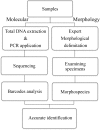
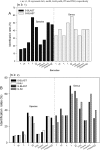
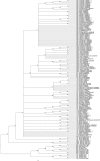
Lauraceae are an important component of tropical and subtropical forests and have major ecological and economic significance. Owing to lack of clear-cut morphological differences between genera and species, this family is an ideal case for testing the efficacy of DNA barcoding in the identification and discrimination of species and genera. In this study, we evaluated five widely recommended plant DNA barcode loci matK, rbcL, trnH-psbA, ITS2 and the entire ITS region for 409 individuals representing 133 species, 12 genera from China. We tested the ability of DNA barcoding to distinguish species and as an alternative tool for correcting species misidentification. We also used the rbcL+matK+trnH-psbA+ITS loci to investigate the phylogenetic relationships of the species examined. Among the gene regions and their combinations, ITS was the most efficient for identifying species (57.5%) and genera (70%). DNA barcoding also had a positive role for correcting species misidentification (10.8%). Furthermore, based on the results of the phylogenetic analyses, Chinese Lauraceae species formed three supported monophyletic clades, with the Cryptocarya group strongly supported (PP = 1.00, BS = 100%) and the clade including the Persea group, Laureae and Cinnamomum also receiving strong support (PP = 1.00, BS = 98%), whereas the Caryodaphnopsis-Neocinnamomum received only moderate support (PP = 1.00 and BS = 85%). This study indicates that molecular barcoding can assist in screening difficult to identify families like Lauraceae, detecting errors of species identification, as well as helping to reconstruct phylogenetic relationships. DNA barcoding can thus help with large-scale biodiversity inventories and rare species conservation by improving accuracy, as well as reducing time and costs associated with species identification.
Conflict of interest statement
Figures


Similar articles
-
Research Progress in Plant Molecular Systematics of Lauraceae.Biology (Basel). 2021 May 1;10(5):391. doi: 10.3390/biology10050391. Biology (Basel). 2021. PMID: 34062846 Free PMC article. Review.
-
Identification of species in the angiosperm family Apiaceae using DNA barcodes.Mol Ecol Resour. 2014 Nov;14(6):1231-8. doi: 10.1111/1755-0998.12262. Epub 2014 May 14. Mol Ecol Resour. 2014. PMID: 24739357
-
DNA barcoding analysis and phylogenetic relationships of tree species in tropical cloud forests.Sci Rep. 2017 Oct 2;7(1):12564. doi: 10.1038/s41598-017-13057-0. Sci Rep. 2017. PMID: 28970548 Free PMC article.
-
Essential Oil Composition and DNA Barcode and Identification of Aniba species (Lauraceae) Growing in the Amazon Region.Molecules. 2021 Mar 29;26(7):1914. doi: 10.3390/molecules26071914. Molecules. 2021. PMID: 33805452 Free PMC article.
-
Traditional System Versus DNA Barcoding in Identification of Bamboo Species: A Systematic Review.Mol Biotechnol. 2021 Aug;63(8):651-675. doi: 10.1007/s12033-021-00337-4. Epub 2021 May 17. Mol Biotechnol. 2021. PMID: 34002354
Cited by
-
DNA Barcoding and Phylogenomic Analysis of the Genus Fritillaria in China Based on Complete Chloroplast Genomes.Front Plant Sci. 2022 Feb 25;13:764255. doi: 10.3389/fpls.2022.764255. eCollection 2022. Front Plant Sci. 2022. PMID: 35283910 Free PMC article.
-
Global advances in phylogeny, taxonomy and biogeography of Lauraceae.Plant Divers. 2025 Apr 7;47(3):341-364. doi: 10.1016/j.pld.2025.04.001. eCollection 2025 May. Plant Divers. 2025. PMID: 40496996 Free PMC article. Review.
-
Research Progress in Plant Molecular Systematics of Lauraceae.Biology (Basel). 2021 May 1;10(5):391. doi: 10.3390/biology10050391. Biology (Basel). 2021. PMID: 34062846 Free PMC article. Review.
-
Phylogenetic analysis of Fritillaria cirrhosa D. Don and its closely related species based on complete chloroplast genomes.PeerJ. 2019 Aug 21;7:e7480. doi: 10.7717/peerj.7480. eCollection 2019. PeerJ. 2019. PMID: 31497389 Free PMC article.
-
Endorsement and phylogenetic analysis of some Fabaceae plants based on DNA barcoding.Mol Biol Rep. 2022 Jun;49(6):5645-5657. doi: 10.1007/s11033-022-07574-z. Epub 2022 Jun 2. Mol Biol Rep. 2022. PMID: 35655052 Free PMC article.
References
-
- Li HW, Li J, Huang PH, Wei FN, Cui HB, van der Werff H. Lauraceae. In: Wu ZY, Raven PH editors. Flora of China, Calycanthaceae–Schisandraceae. Vol.7 Beijing, and St. Louis, Missouri: Science Press and Missouri Botanical Garden Press; 2008. pp. 102–254.
-
- Yang Y, Liu B. Species catalogue of Lauraceae in China: problems and perspectives. Biod Sci. 2015; 23(2): 232–236.
-
- van der Werff H, Richter HG. Toward an improved classification of Lauraceae. Ann Missouri Bot Gard. 1996; 83: 409–418.
-
- Van der Werff H. An annotated key to the genera of Lauraceae in the Flora Malesiana Region. Blumea. 2001; 46: 125–140.
-
- Rohwer JG. Lauraceae In: Kubitzki K, Rohwer JG, Brittrich V editors. The families and genera of vascular plants. Vol.2 Berlin: Springer-Verlag; 1993. pp. 366–391.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

