Lipidomics by ultrahigh performance liquid chromatography-high resolution mass spectrometry and its application to complex biological samples
- PMID: 28415015
- PMCID: PMC6013032
- DOI: 10.1016/j.jchromb.2017.03.027
Lipidomics by ultrahigh performance liquid chromatography-high resolution mass spectrometry and its application to complex biological samples
Abstract
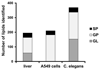
An improved approach for selective and sensitive identification and quantitation of lipid molecular species using reversed phase chromatography coupled to high resolution mass spectrometry was developed. The method is applicable to a wide variety of biological matrices using a simple liquid-liquid extraction procedure. Together, this approach combines multiple selectivity criteria: Reversed phase chromatography separates lipids according to their acyl chain length and degree of unsaturation and is capable of resolving positional isomers of lysophospholipids, as well as structural isomers of diacyl phospholipids and glycerolipids. Orbitrap mass spectrometry delivers the elemental composition of both positive and negative ions with high mass accuracy. Finally, automatically generated tandem mass spectra provide structural insight into numerous glycerolipids, phospholipids, and sphingolipids within a single run. Calibration showed linearity ranges of more than four orders of magnitude, good values for accuracy and precision at biologically relevant concentration levels, and limits of quantitation of a few femtomoles on column. Hundreds of lipid molecular species were detected and quantified in three different biological matrices, which cover well the wide variety and complexity of various model organisms in lipidomic research. Together with a software package, this method is a prime choice for global lipidomic analysis of even the most complex biological samples.
Keywords: High performance liquid chromatography; High-resolution mass spectrometry; Lipid structural isomers; Quantitative lipidomics.
Copyright © 2017 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of Interest
All authors declare no conflict of interest.
Figures


References
-
- Fahy E, Subramaniam S, Brown HA, Glass CK, Merrill AH, Murphy RC, et al. A comprehensive classification system for lipids. J Lipid Res. 2005;46:839–61. - PubMed
-
- Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610. - PubMed
-
- Ejsing CS, Duchoslav E, Sampaio J, Simons K, Bonner R, Thiele C, et al. Automated Identification and Quantification of Glycerophospholipid Molecular Species by Multiple Precursor Ion Scanning. Anal Chem. 2006;78:6202–14. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

