The N-terminal polypeptide derived from viral macrophage inflammatory protein II reverses breast cancer epithelial-to-mesenchymal transition via a PDGFRα-dependent mechanism
- PMID: 28415580
- PMCID: PMC5514921
- DOI: 10.18632/oncotarget.16394
The N-terminal polypeptide derived from viral macrophage inflammatory protein II reverses breast cancer epithelial-to-mesenchymal transition via a PDGFRα-dependent mechanism
Abstract
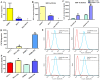
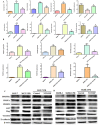
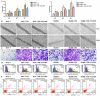
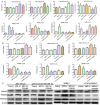
NT21MP, a 21-residue peptide derived from the viral macrophage inflammatory protein II, competed effectively with the natural ligand of CXC chemokine receptor 4 (CXCR4), stromal cell-derived factor 1-alpha, to induce apoptosis and inhibit growth in breast cancer. Its role in tumor epithelial-to-mesenchymal transition (EMT) regulation remains unknown. In this study, we evaluated the reversal of EMT upon NT21MP treatment and examined its role in the inhibition of EMT in breast cancer. The parental cells of breast cancer (SKBR-3 and MCF-7) and paclitaxel-resistant (SKBR-3 PR and MCF-7 PR) cells were studied in vitro and in combined immunodeficient mice. The mice injected with SKBR-3 PR cells were treated with NT21MP through the tail vein or intraperitoneally with paclitaxel or saline. Sections from tumors were evaluated for tumor weight and EMT markers based on Western blot. In vitro, the effects of NT21MP, CXCR4 and PDGFRα on tumor EMT were assessed by relative quantitative real-time reverse transcription-polymerase chain reaction, western blot and biological activity in breast cancer cell lines expressing high or low levels of CXCR4. Our results illustrated that NT21MP could reverse the phenotype of EMT in paclitaxel-resistant cells. Furthermore, we found that NT21MP governed PR-mediated EMT partly due to controlling platelet-derived growth factors A and B (PDGFA and PDGFB) and their receptor (PDGFRα). More importantly, NT21MP down-regulated AKT and ERK1/2 activity, which were activated by PDGFRα, and eventually reversed the EMT. Together, these results indicated that CXCR4 overexpression drives acquired paclitaxel resistance, partly by activating the PDGFA and PDGFB/PDGFRα autocrine signaling loops that activate AKT and ERK1/2. Inhibition of the oncogenic EMT process by targeting CXCR4/PDGFRα-mediated pathways using NT21MP may provide a novel therapeutic approach towards breast cancer.
Keywords: PDGFRα; breast cancer; epithelial-mesenchymal transition; paclitaxel; viral macrophage inflammatory protein II.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures




References
-
- Fan L, Strasser-Weippl K, Li JJ, St Louis J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM, Goss PE. Breast cancer in China. Lancet Oncol. 2014;15:279–289. - PubMed
-
- Siegal R, Naishadham D, Jemal A. Cancer Statistics. CA Cancer J Clin. 2013;2013(63):11–30. - PubMed
-
- NCCN Clinical . Practice Guidelines in Oncology (NCCN Guidelines), Breast Cancer. Version 3.2014 www.NCCN.org/patients.
-
- De Lena M, Latorre A, Calabrese P, Catino A, Lorusso V, Mazzei A, Aloe A. High efficacy of paclitaxel and doxorubicin as first-line therapy in advanced breast cancer: a phase I-II study. J Chemother. 2000;12:367–373. - PubMed
-
- Sabbah M, Emami S, Redeuilh G, Julien S, Prévost G, Zimber A, Ouelaa R, Bracke M, De Wever O, Gespach C. Molecular signature and therapeutic perspective of the epithelial-to-mesenchymal transitions in epithelial cancers. Drug Resist Updat. 2008;11:123–151. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

