A(H1N1)pdm09 influenza infection: vaccine inefficiency
- PMID: 28415629
- PMCID: PMC5464833
- DOI: 10.18632/oncotarget.16459
A(H1N1)pdm09 influenza infection: vaccine inefficiency
Abstract
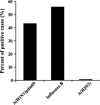
The last influenza pandemic, caused by the swine A(H1N1)pdm09 influenza virus, began in North America at 2009. Since then, the World Health Organization (WHO) recommended integration of the swine-based virus A/California/07/2009 strain in yearly vaccinations. Yet, infections with A(H1N1)pdm09 have continued in subsequent years. The reasons for this are currently unknown. During the 2015-2016 influenza season, we noted an increased prevalence of A(H1N1)pdm09 influenza virus infection in Israel. Our phylogenetic analysis indicated that the circulating A(H1N1)pdm09 strains belonged to 6B.1 and 6B.2 clades and differed from the vaccinating strain, with approximately 18 amino acid differences found between the circulating strains and the immunizing A/California/07/2009 strain. Hemmaglutination inhibition (HI) assays demonstrated higher antibodies titer against the A/California/07/2009 vaccinating strain as compared to the circulating Israeli strains. We thus suggest that the current vaccination was not sufficiently effective and propose inclusion of the current circulating A(H1N1)pdm09 influenza viruses in the annual vaccine composition.
Keywords: Clade 6B; H1N1; influenza A; vaccine.
Conflict of interest statement
None.
Figures






References
-
- Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, Cox NJ, Fukuda K. Influenza-associated hospitalizations in the United States. JAMA. 2004;292:1333–1340. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

