The combined effect of USP7 inhibitors and PARP inhibitors in hormone-sensitive and castration-resistant prostate cancer cells
- PMID: 28415632
- PMCID: PMC5458250
- DOI: 10.18632/oncotarget.16463
The combined effect of USP7 inhibitors and PARP inhibitors in hormone-sensitive and castration-resistant prostate cancer cells
Abstract
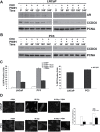
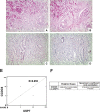
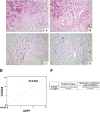
Purpose of the study: Reduced levels of the tumor suppressor protein CCDC6 sensitize cancer cells to the treatment with PARP-inhibitors. The turnover of CCDC6 protein is regulated by the de-ubiquitinase USP7, which also controls the androgen receptor (AR) stability. Here, we correlated the expression levels of CCDC6 and USP7 proteins in primary prostate cancers (PC). Moreover, we tested the efficacy of the USP7 inhibitors, in combination with PARP-inhibitors as a novel therapeutic option in advanced prostate cancer.Experimental techniques: PC cells were exposed to USP7 inhibitor, P5091, together with cycloheximide, to investigate the turnover of the USP7 substrates, AR and CCDC6. As outcome of the AR downregulation, transcription targets of AR and its variant V7 were examined by qPCR. As a result of CCDC6 degradation, the induction of PARP inhibitors sensitivity was evaluated by analyzing PC cells viability and foci formation. We scored and correlated CCDC6 and USP7 expression levels in a prostate cancer tissue microarray (TMA).
Results: P5091 accelerated the degradation of AR and V7 isoform affecting PSA, UBE2C, CDC20 transcription and PC cells proliferation. Moreover, P5091 accelerated the degradation of CCDC6 sensitizing the cells to PARP-inhibitors, that acted sinergistically with genotoxic agents. The immunohistochemical analysis of both CCDC6 and USP7 proteins exhibited significant correlation for the intensity of staining (p ≤ 0.05).Data interpretation: Thus, CCDC6 and USP7 represent predictive markers for the combined treatment of the USP7-inhibitors and PARP-inhibitors in advanced prostate cancer.
Keywords: ARFL and V7; CCDC6; P5091; USP7; olaparib.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
CCDC6 and USP7 expression levels suggest novel treatment options in high-grade urothelial bladder cancer.J Exp Clin Cancer Res. 2019 Feb 20;38(1):90. doi: 10.1186/s13046-019-1087-1. J Exp Clin Cancer Res. 2019. PMID: 30786932 Free PMC article.
-
USP7 inhibitors, downregulating CCDC6, sensitize lung neuroendocrine cancer cells to PARP-inhibitor drugs.Lung Cancer. 2017 May;107:41-49. doi: 10.1016/j.lungcan.2016.06.015. Epub 2016 Jun 17. Lung Cancer. 2017. PMID: 27372520
-
Analysis of CCDC6 as a novel biomarker for the clinical use of PARP1 inhibitors in malignant pleural mesothelioma.Lung Cancer. 2019 Sep;135:56-65. doi: 10.1016/j.lungcan.2019.07.011. Epub 2019 Jul 13. Lung Cancer. 2019. PMID: 31447003
-
Identification of Novel Biomarkers of Homologous Recombination Defect in DNA Repair to Predict Sensitivity of Prostate Cancer Cells to PARP-Inhibitors.Int J Mol Sci. 2019 Jun 25;20(12):3100. doi: 10.3390/ijms20123100. Int J Mol Sci. 2019. PMID: 31242618 Free PMC article. Review.
-
Clinical Development of PARP Inhibitors in Treating Metastatic Castration-Resistant Prostate Cancer.Cells. 2019 Aug 9;8(8):860. doi: 10.3390/cells8080860. Cells. 2019. PMID: 31404966 Free PMC article. Review.
Cited by
-
Highlights in USP7 inhibitors for cancer treatment.Front Chem. 2022 Sep 15;10:1005727. doi: 10.3389/fchem.2022.1005727. eCollection 2022. Front Chem. 2022. PMID: 36186590 Free PMC article. Review.
-
Response prediction biomarkers and drug combinations of PARP inhibitors in prostate cancer.Acta Pharmacol Sin. 2021 Dec;42(12):1970-1980. doi: 10.1038/s41401-020-00604-1. Epub 2021 Feb 15. Acta Pharmacol Sin. 2021. PMID: 33589795 Free PMC article. Review.
-
Drug resistance mechanisms and treatment strategies mediated by Ubiquitin-Specific Proteases (USPs) in cancers: new directions and therapeutic options.Mol Cancer. 2024 May 3;23(1):88. doi: 10.1186/s12943-024-02005-y. Mol Cancer. 2024. PMID: 38702734 Free PMC article. Review.
-
USP7 promotes temozolomide resistance by stabilizing MGMT in glioblastoma.Cell Death Dis. 2025 Aug 20;16(1):631. doi: 10.1038/s41419-025-07969-3. Cell Death Dis. 2025. PMID: 40835840 Free PMC article.
-
Ubiquitin-specific protease 7 as a potential therapeutic target in dogs with hematopoietic malignancies.J Vet Intern Med. 2021 Mar;35(2):1041-1051. doi: 10.1111/jvim.16082. Epub 2021 Mar 2. J Vet Intern Med. 2021. PMID: 33650720 Free PMC article.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. - PubMed
-
- Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–1403. - PubMed
-
- Pienta KJ, Bradley D. Mechanisms underlying the development of androgen-independent prostate cancer. Clin Cancer Res. 2006;12:1665–1671. - PubMed
-
- Holzbeierlein J, Lal P, LaTulippe E, Smith A, Satagopan J, Zhang L, Ryan C, Smith S, Scher H, Scardino P, Reuter V, Gerald WL. Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. Am J Pathol. 2004;164:217–27. - PMC - PubMed
-
- Debes JD, Tindall DJ. Mechanisms of androgen-refractory prostate cancer. N Engl J Med. 2004;351:1488–90. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

