The associations between the MAPT polymorphisms and Alzheimer's disease risk: a meta-analysis
- PMID: 28415654
- PMCID: PMC5522165
- DOI: 10.18632/oncotarget.16490
The associations between the MAPT polymorphisms and Alzheimer's disease risk: a meta-analysis
Abstract
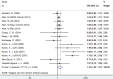
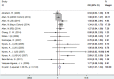
Published studies revealed that the microtubule-associated protein tau (MAPT) gene polymorphisms increased Alzheimer's disease (AD) risk; the associations of 4 single nucleotide polymorphisms (SNPs, rs242557G/A, rs2471738C/T, rs3785883G/A and rs1467967A/G) of the MAPT gene with AD risk, however, remain inconclusive. Here, we conducted a meta-analysis to investigate the relationship between the MAPT SNPs and AD risk. A significant association of SNP rs242557 with AD risk was found in a dominant [odds ratio (OR) = 1.05, 95% confidence interval (CI) = 1.01, 1.10, P = 0.025] genetic model, and a suggestive association in an allelic (OR = 1.03, 95% CI = 1.00, 1.06, P = 0.078). When APOE epsilon 4 carrier status was included in stratified analysis, this association was even stronger (allelic model for the APOE epsilon 4 positive individuals: OR = 1.24, 95% CI = 1.08, 1.43, P = 0.003). Furthermore, a significant association of SNP rs2471738 with AD risk was found under all the four models (allelic: OR = 1.11, 95% CI = 1.01, 1.20, P = 0.021; dominant: OR = 1.10, 95% CI = 1.00, 1.21, P = 0.046; recessive: OR = 1.18, 95% CI = 1.05, 1.32, P = 0.004; additive: OR = 1.20, 95% CI = 1.07, 1.34, P = 0.002) models. However, pooled results suggest that the neither rs3785883 nor rs1467967 is associated with AD risk under all the four genetic models. In summary, our study provides further evidence of the associations of the MAPT SNPs with AD risk.
Keywords: Alzheimer’s disease; meta-analysis; microtubule-associated protein tau; single nucleotide polymorphisms.
Conflict of interest statement
The authors declare that they have no conflict of interests.
Figures







Similar articles
-
Association of microtubule-associated protein tau gene polymorphisms with the risk of sporadic Alzheimer's disease: a meta-analysis.Int J Neurosci. 2018 Jun;128(6):577-585. doi: 10.1080/00207454.2017.1400972. Int J Neurosci. 2018. PMID: 29098924
-
Meta-analysis of the association between variants in MAPT and neurodegenerative diseases.Oncotarget. 2017 Jul 4;8(27):44994-45007. doi: 10.18632/oncotarget.16690. Oncotarget. 2017. PMID: 28402959 Free PMC article. Review.
-
Association of MAPT haplotype-tagging polymorphisms with cerebrospinal fluid biomarkers of Alzheimer's disease: A preliminary study in a Croatian cohort.Brain Behav. 2018 Nov;8(11):e01128. doi: 10.1002/brb3.1128. Epub 2018 Oct 17. Brain Behav. 2018. PMID: 30329219 Free PMC article.
-
Structural study of the microtubule-associated protein tau locus of Alzheimer's disease in Taiwan.Biomed J. 2014 May-Jun;37(3):127-32. doi: 10.4103/2319-4170.117891. Biomed J. 2014. PMID: 24923570
-
Influence of four polymorphisms in ABCA1 and PTGS2 genes on risk of Alzheimer's disease: a meta-analysis.Neurol Sci. 2016 Aug;37(8):1209-20. doi: 10.1007/s10072-016-2579-9. Epub 2016 May 23. Neurol Sci. 2016. PMID: 27215623 Review.
Cited by
-
Preliminary exploration of the co-regulation of Alzheimer's disease pathogenic genes by microRNAs and transcription factors.Front Aging Neurosci. 2022 Dec 6;14:1069606. doi: 10.3389/fnagi.2022.1069606. eCollection 2022. Front Aging Neurosci. 2022. PMID: 36561136 Free PMC article.
-
Non-invasive and high-throughput interrogation of exon-specific isoform expression.Nat Cell Biol. 2021 Jun;23(6):652-663. doi: 10.1038/s41556-021-00678-x. Epub 2021 Jun 3. Nat Cell Biol. 2021. PMID: 34083785 Free PMC article.
-
Validation of Enhancer Regions in Primary Human Neural Progenitor Cells using Capture STARR-seq.bioRxiv [Preprint]. 2024 Mar 18:2024.03.14.585066. doi: 10.1101/2024.03.14.585066. bioRxiv. 2024. Update in: Genome Res. 2025 Aug 1;35(8):1887-1901. doi: 10.1101/gr.279584.124. PMID: 38562832 Free PMC article. Updated. Preprint.
-
Genome-wide meta-analysis identifies 127 open-angle glaucoma loci with consistent effect across ancestries.Nat Commun. 2021 Feb 24;12(1):1258. doi: 10.1038/s41467-020-20851-4. Nat Commun. 2021. PMID: 33627673 Free PMC article.
-
Potential common pathogenesis of several neurodegenerative diseases.Neural Regen Res. 2026 Mar 1;21(3):972-988. doi: 10.4103/NRR.NRR-D-24-01054. Epub 2025 May 30. Neural Regen Res. 2026. PMID: 40522761 Free PMC article.
References
-
- Giacobini E, Gold G. Alzheimer disease therapy—moving from amyloid-beta to tau. Nat Rev Neurol. 2013;9:677–686. - PubMed
-
- Scheltens P, Blennow K, Breteler MM, de Strooper B, Frisoni GB, Salloway S, Van der Flier WM. Alzheimer’s disease. The Lancet. 2016;388:505–517. - PubMed
-
- Spillantini MG, Goedert M. Tau pathology and neurodegeneration. Lancet Neurol. 2013;12:609–622. - PubMed
-
- Liu QY, Yu JT, Miao D, Ma XY, Wang HF, Wang W, Tan L. An exploratory study on STX6, MOBP, MAPT, and EIF2AK3 and late-onset Alzheimer's disease. Neurobiol Aging. 2013;34:1513–1519. - PubMed
-
- Laws SM, Friedrich P, Diehl-Schmid J, Muller J, Eisele T, Bauml J, Forstl H, Kurz A, Riemenschneider M. Fine mapping of the MAPT locus using quantitative trait analysis identifies possible causal variants in Alzheimer’s disease. Mol Psychiatry. 2007;12:510–517. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

