A feasibility study of returning clinically actionable somatic genomic alterations identified in a research laboratory
- PMID: 28415679
- PMCID: PMC5522029
- DOI: 10.18632/oncotarget.16018
A feasibility study of returning clinically actionable somatic genomic alterations identified in a research laboratory
Erratum in
-
Correction: A feasibility study of returning clinically actionable somatic genomic alterations identified in a research laboratory.Oncotarget. 2019 Aug 27;10(50):5254. doi: 10.18632/oncotarget.27176. eCollection 2019 Aug 27. Oncotarget. 2019. PMID: 31497255 Free PMC article.
Abstract
Purpose: Molecular profiling performed in the research setting usually does not benefit the patients that donate their tissues. Through a prospective protocol, we sought to determine the feasibility and utility of performing broad genomic testing in the research laboratory for discovery, and the utility of giving treating physicians access to research data, with the option of validating actionable alterations in the CLIA environment.
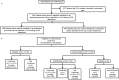
Experimental design: 1200 patients with advanced cancer underwent characterization of their tumors with high depth hybrid capture sequencing of 201 genes in the research setting. Tumors were also tested in the CLIA laboratory, with a standardized hotspot mutation analysis on an 11, 46 or 50 gene platform.
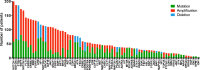
Results: 527 patients (44%) had at least one likely somatic mutation detected in an actionable gene using hotspot testing. With the 201 gene panel, 945 patients (79%) had at least one alteration in a potentially actionable gene that was undetected with the more limited CLIA panel testing. Sixty-four genomic alterations identified on the research panel were subsequently tested using an orthogonal CLIA assay. Of 16 mutations tested in the CLIA environment, 12 (75%) were confirmed. Twenty-five (52%) of 48 copy number alterations were confirmed. Nine (26.5%) of 34 patients with confirmed results received genotype-matched therapy. Seven of these patients were enrolled onto genotype-matched targeted therapy trials.
Conclusion: Expanded cancer gene sequencing identifies more actionable genomic alterations. The option of CLIA validating research results can provide alternative targets for personalized cancer therapy.
Keywords: CLIA; clinical trial; genomics; precision medicine; somatic mutation.
Conflict of interest statement
The authors declare no potential conflicts of interest and have no financial disclosures.
Figures

References
-
- Robson ME, Bradbury AR, Arun B, Domchek SM, Ford JM, Hampel HL, Lipkin SM, Syngal S, Wollins DS, Lindor NM. American Society of Clinical Oncology Policy Statement Update: Genetic and Genomic Testing for Cancer Susceptibility. J Clin Oncol. 2015;33:3660–67. - PubMed
-
- Meric-Bernstam F, Brusco L, Shaw K, Horombe C, Kopetz S, Davies MA, Routbort M, Piha-Paul SA, Janku F, Ueno N, Hong D, De Groot J, Ravi V, et al. Feasibility of Large-Scale Genomic Testing to Facilitate Enrollment Onto Genomically Matched Clinical Trials. J Clin Oncol. 2015;33:2753–62. - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

