Drosophila melanogaster retrotransposon and inverted repeat-derived endogenous siRNAs are differentially processed in distinct cellular locations
- PMID: 28415970
- PMCID: PMC5392987
- DOI: 10.1186/s12864-017-3692-8
Drosophila melanogaster retrotransposon and inverted repeat-derived endogenous siRNAs are differentially processed in distinct cellular locations
Abstract
Background: Endogenous small interfering (esi)RNAs repress mRNA levels and retrotransposon mobility in Drosophila somatic cells by poorly understood mechanisms. 21 nucleotide esiRNAs are primarily generated from retrotransposons and two inverted repeat (hairpin) loci in Drosophila culture cells in a Dicer2 dependent manner. Additionally, proteins involved in 3' end processing, such as Symplekin, CPSF73 and CPSR100, have been recently implicated in the esiRNA pathway.
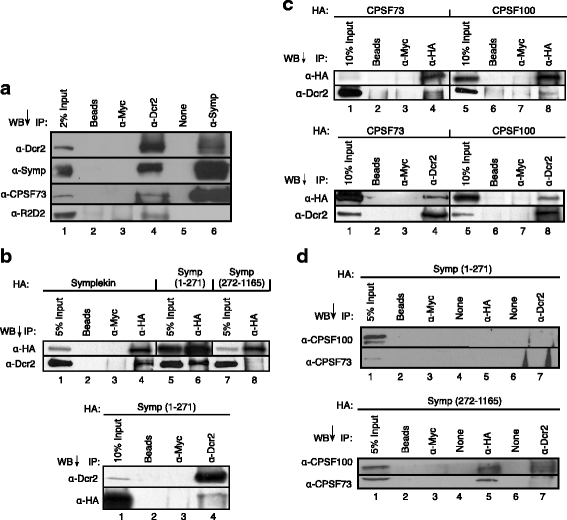
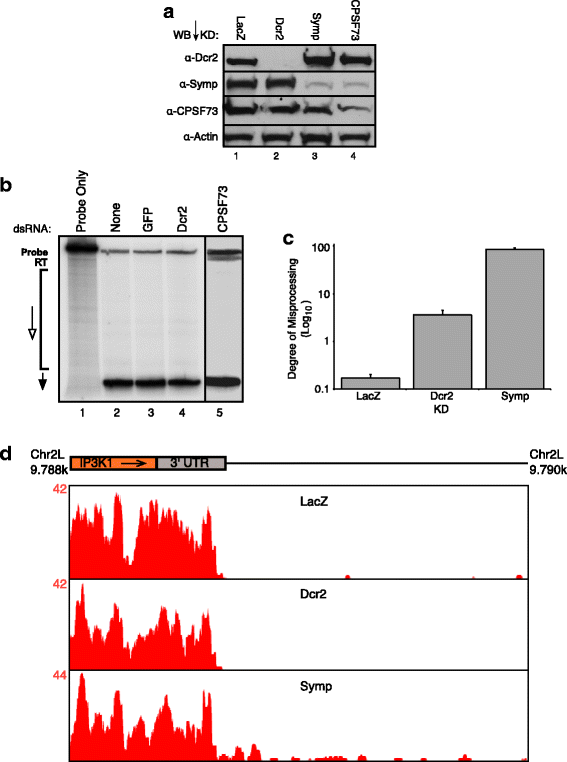
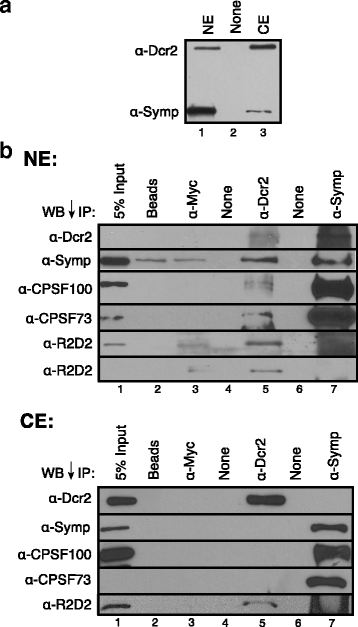
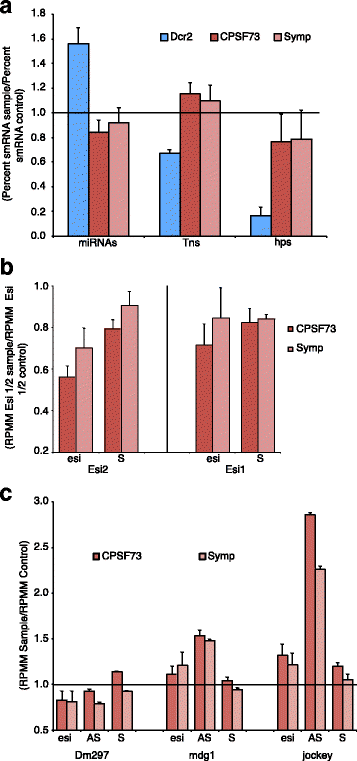
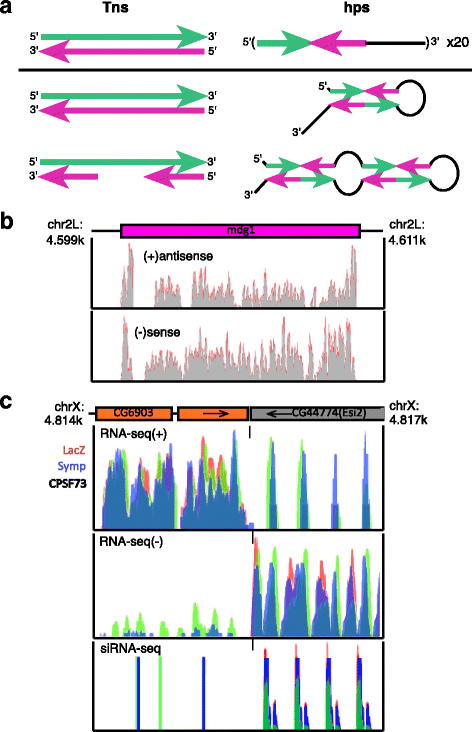
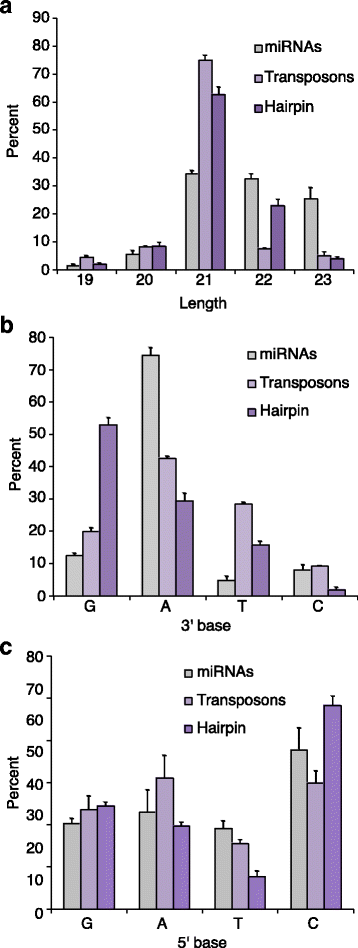
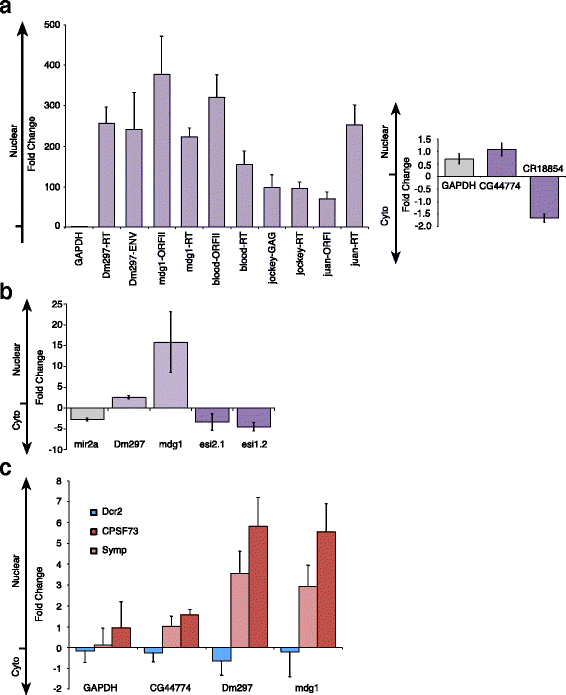
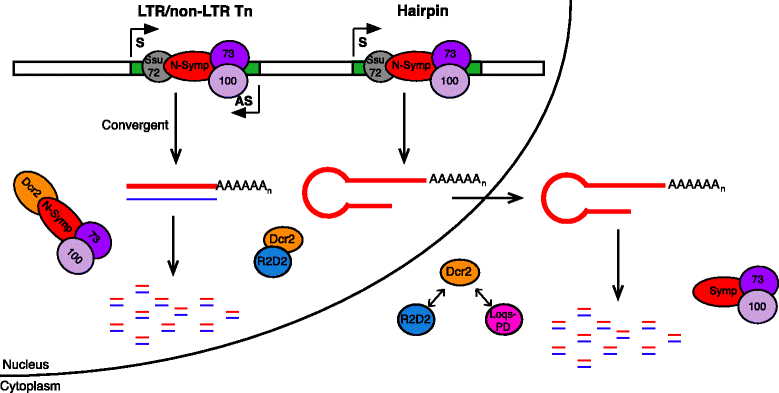
Results: Here we present evidence of overlap between two essential RNA metabolic pathways: esiRNA biogenesis and mRNA 3' end processing. We have identified a nucleus-specific interaction between the essential esiRNA cleavage enzyme Dicer2 (Dcr2) and Symplekin, a component of the core cleavage complex (CCC) required for 3' end processing of all eukaryotic mRNAs. This interaction is mediated by the N-terminal 271 amino acids of Symplekin; CCC factors CPSF73 and CPSF100 do not contact Dcr2. While Dcr2 binds the CCC, Dcr2 knockdown does not affect mRNA 3' end formation. RNAi-depletion of CCC components Symplekin and CPSF73 causes perturbations in esiRNA abundance that correlate with fluctuations in retrotransposon and hairpin esiRNA precursor levels. We also discovered that esiRNAs generated from retrotransposons and hairpins have distinct physical characteristics including a higher predominance of 22 nucleotide hairpin-derived esiRNAs and differences in 3' and 5' base preference. Additionally, retrotransposon precursors and derived esiRNAs are highly enriched in the nucleus while hairpins and hairpin derived esiRNAs are predominantly cytoplasmic similar to canonical mRNAs. RNAi-depletion of either CPSF73 or Symplekin results in nuclear retention of both hairpin and retrotransposon precursors suggesting that polyadenylation indirectly affects cellular localization of Dcr2 substrates.
Conclusions: Together, these observations support a novel mechanism in which differences in localization of esiRNA precursors impacts esiRNA biogenesis. Hairpin-derived esiRNAs are generated in the cytoplasm independent of Dcr2-Symplekin interactions, while retrotransposons are processed in the nucleus.
Keywords: CPSF73; Core cleavage complex; Dicer2; Endogenous small interfering RNA biogenesis; Symplekin; mRNA 3’ end processing.
Figures
Similar articles
-
In vivo characterization of the Drosophila mRNA 3' end processing core cleavage complex.RNA. 2015 Aug;21(8):1404-18. doi: 10.1261/rna.049551.115. Epub 2015 Jun 16. RNA. 2015. PMID: 26081560 Free PMC article.
-
A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs.Mol Cell. 2009 May 15;34(3):322-32. doi: 10.1016/j.molcel.2009.04.024. Mol Cell. 2009. PMID: 19450530 Free PMC article.
-
Antisense Transcription of Retrotransposons in Drosophila: An Origin of Endogenous Small Interfering RNA Precursors.Genetics. 2016 Jan;202(1):107-21. doi: 10.1534/genetics.115.177196. Epub 2015 Nov 3. Genetics. 2016. PMID: 26534950 Free PMC article.
-
How selfish retrotransposons are silenced in Drosophila germline and somatic cells.FEBS Lett. 2008 Jul 23;582(17):2473-8. doi: 10.1016/j.febslet.2008.06.018. Epub 2008 Jun 20. FEBS Lett. 2008. PMID: 18572018 Review.
-
Formation of the 3' end of histone mRNA: getting closer to the end.Gene. 2007 Jul 15;396(2):373-90. doi: 10.1016/j.gene.2007.04.021. Epub 2007 May 4. Gene. 2007. PMID: 17531405 Free PMC article. Review.
Cited by
-
Modulation of diverse biological processes by CPSF, the master regulator of mRNA 3' ends.RNA. 2024 Aug 16;30(9):1122-1140. doi: 10.1261/rna.080108.124. RNA. 2024. PMID: 38986572 Free PMC article. Review.
-
In vitro studies provide insight into effects of Dicer-2 helicase mutations in Drosophila melanogaster.RNA. 2020 Dec;26(12):1847-1861. doi: 10.1261/rna.077289.120. Epub 2020 Aug 25. RNA. 2020. PMID: 32843367 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

