The deubiquitinating enzyme Usp14 allosterically inhibits multiple proteasomal activities and ubiquitin-independent proteolysis
- PMID: 28416611
- PMCID: PMC5465503
- DOI: 10.1074/jbc.M116.763128
The deubiquitinating enzyme Usp14 allosterically inhibits multiple proteasomal activities and ubiquitin-independent proteolysis
Abstract
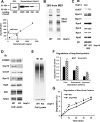
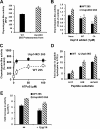
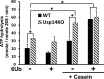
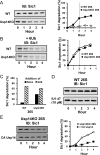
The proteasome-associated deubiquitinating enzyme Usp14/Ubp6 inhibits protein degradation by catalyzing substrate deubiquitination and by poorly understood allosteric actions. However, upon binding a ubiquitin chain, Usp14 enhances proteasomal degradation by stimulating ATP and peptide degradation. These studies were undertaken to clarify these seemingly opposite regulatory roles of Usp14 and their importance. To learn how the presence of Usp14 on 26S proteasomes influences its different activities, we compared enzymatic and regulatory properties of 26S proteasomes purified from wild-type mouse embryonic fibroblast cells and those lacking Usp14. The proteasomes lacking Usp14 had higher basal peptidase activity than WT 26S, and this activity was stimulated to a greater extent by adenosine 5'-O-(thiotriphosphate) (ATPγS) than with WT particles. These differences were clear even though Usp14 is present on only a minor fraction (30-40%) of the 26S in WT mouse embryonic fibroblast cells. Addition of purified Usp14 to the WT and Usp14-defficient proteasomes reduced both their basal peptidase activity and the stimulation by ATPγS. Usp14 inhibits these processes allosterically because a catalytically inactive Usp14 mutant also inhibited them. Proteasomes lacking Usp14 also exhibited greater deubiquitinating activity by Rpn11 and greater basal ATPase activity than WT particles. ATP hydrolysis by WT proteasomes is activated if they bind a ubiquitinated protein, which is loosely folded. Surprisingly, proteasomes lacking Usp14 could be activated by such proteins even without a ubiquitin chain present. Furthermore, proteasomes lacking Usp14 are much more active in degrading non-ubiquitinated proteins (e.g. Sic1) than WT particles. Thus, without a ubiquitinated substrate present, Usp14 suppresses multiple proteasomal activities, especially basal ATP consumption and degradation of non-ubiquitinated proteins. These allosteric effects thus reduce ATP hydrolysis by inactive proteasomes and nonspecific proteolysis and enhance proteasomal specificity for ubiquitinated proteins.
Keywords: ATPase; Usp14; allosteric regulation; deubiquitylation (deubiquitination); gate opening; peptidase; proteasome; protein degradation.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

Similar articles
-
UBL domain of Usp14 and other proteins stimulates proteasome activities and protein degradation in cells.Proc Natl Acad Sci U S A. 2018 Dec 11;115(50):E11642-E11650. doi: 10.1073/pnas.1808731115. Epub 2018 Nov 28. Proc Natl Acad Sci U S A. 2018. PMID: 30487212 Free PMC article.
-
Ubiquitinated proteins promote the association of proteasomes with the deubiquitinating enzyme Usp14 and the ubiquitin ligase Ube3c.Proc Natl Acad Sci U S A. 2017 Apr 25;114(17):E3404-E3413. doi: 10.1073/pnas.1701734114. Epub 2017 Apr 10. Proc Natl Acad Sci U S A. 2017. PMID: 28396413 Free PMC article.
-
In vitro analysis of proteasome-associated USP14 activity for substrate degradation and deubiquitylation.Methods Enzymol. 2019;619:249-268. doi: 10.1016/bs.mie.2018.12.028. Epub 2019 Feb 1. Methods Enzymol. 2019. PMID: 30910023
-
Deubiquitination Reactions on the Proteasome for Proteasome Versatility.Int J Mol Sci. 2020 Jul 27;21(15):5312. doi: 10.3390/ijms21155312. Int J Mol Sci. 2020. PMID: 32726943 Free PMC article. Review.
-
Trimming of ubiquitin chains by proteasome-associated deubiquitinating enzymes.Mol Cell Proteomics. 2011 May;10(5):R110.003871. doi: 10.1074/mcp.R110.003871. Epub 2010 Sep 7. Mol Cell Proteomics. 2011. PMID: 20823120 Free PMC article. Review.
Cited by
-
Advances in Proteasome Enhancement by Small Molecules.Biomolecules. 2021 Nov 30;11(12):1789. doi: 10.3390/biom11121789. Biomolecules. 2021. PMID: 34944433 Free PMC article. Review.
-
FAT10 and NUB1L cooperate to activate the 26S proteasome.Life Sci Alliance. 2023 May 15;6(8):e202201463. doi: 10.26508/lsa.202201463. Print 2023 Aug. Life Sci Alliance. 2023. PMID: 37188463 Free PMC article.
-
Inactive USP14 and inactive UCHL5 cause accumulation of distinct ubiquitinated proteins in mammalian cells.PLoS One. 2019 Nov 8;14(11):e0225145. doi: 10.1371/journal.pone.0225145. eCollection 2019. PLoS One. 2019. PMID: 31703099 Free PMC article.
-
Meddling with Fate: The Proteasomal Deubiquitinating Enzymes.J Mol Biol. 2017 Nov 10;429(22):3525-3545. doi: 10.1016/j.jmb.2017.09.015. Epub 2017 Oct 5. J Mol Biol. 2017. PMID: 28988953 Free PMC article. Review.
-
Ubiquitin-proteasome system as a target for anticancer treatment-an update.Arch Pharm Res. 2023 Jul;46(7):573-597. doi: 10.1007/s12272-023-01455-0. Epub 2023 Aug 5. Arch Pharm Res. 2023. PMID: 37541992 Review.
References
-
- Smith D. M., Benaroudj N., and Goldberg A. (2006) Proteasomes and their associated ATPases: a destructive combination. J. Struct. Biol. 156, 72–83 - PubMed
-
- Kisselev A. F., Garcia-Calvo M., Overkleeft H. S., Peterson E., Pennington M. W., Ploegh H. L., Thornberry N. A., and Goldberg A. L. (2003) The caspase-like sites of proteasomes, their substrate specificity, new inhibitors and substrates, and allosteric interactions with the trypsin-like sites. J. Biol. Chem. 278, 35869–35877 - PubMed
-
- Groll M., and Huber R. (2003) Substrate access and processing by the 20S proteasome core particle. Int. J. Biochem. Cell Biol. 35, 606–616 - PubMed
-
- Groll M., Bajorek M., Köhler A., Moroder L., Rubin D. M., Huber R., Glickman M. H., and Finley D. (2000) A gated channel into the proteasome core particle. Nat. Struct. Biol. 7, 1062–1067 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

