Hypoxia-inducible factor 1α activates insulin-induced gene 2 (Insig-2) transcription for degradation of 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase in the liver
- PMID: 28416613
- PMCID: PMC5454117
- DOI: 10.1074/jbc.M117.788562
Hypoxia-inducible factor 1α activates insulin-induced gene 2 (Insig-2) transcription for degradation of 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase in the liver
Abstract
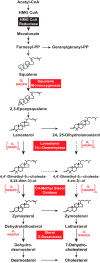
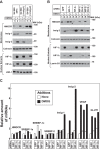
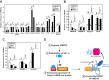
Cholesterol synthesis is a highly oxygen-consuming process. As such, oxygen deprivation (hypoxia) limits cholesterol synthesis through incompletely understood mechanisms mediated by the oxygen-sensitive transcription factor hypoxia-inducible factor 1α (HIF-1α). We show here that HIF-1α links pathways for oxygen sensing and feedback control of cholesterol synthesis in human fibroblasts by directly activating transcription of the INSIG-2 gene. Insig-2 is one of two endoplasmic reticulum membrane proteins that inhibit cholesterol synthesis by mediating sterol-induced ubiquitination and subsequent endoplasmic reticulum-associated degradation of the rate-limiting enzyme in the pathway, HMG-CoA reductase (HMGCR). Consistent with the results in cultured cells, hepatic levels of Insig-2 mRNA were enhanced in mouse models of hypoxia. Moreover, pharmacologic stabilization of HIF-1α in the liver stimulated HMGCR degradation via a reaction that requires the protein's prior ubiquitination and the presence of the Insig-2 protein. In summary, our results show that HIF-1α activates INSIG-2 transcription, leading to accumulation of Insig-2 protein, which binds to HMGCR and triggers its accelerated ubiquitination and degradation. These results indicate that HIF-mediated induction of Insig-2 and degradation of HMGCR are physiologically relevant events that guard against wasteful oxygen consumption and inappropriate cell growth during hypoxia.
Keywords: ER-associated degradation; cholesterol metabolism; endoplasmic reticulum (ER); hypoxia; isoprenoid.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
Figures




Similar articles
-
Hypoxia stimulates degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase through accumulation of lanosterol and hypoxia-inducible factor-mediated induction of insigs.J Biol Chem. 2007 Sep 14;282(37):27436-27446. doi: 10.1074/jbc.M704976200. Epub 2007 Jul 16. J Biol Chem. 2007. PMID: 17635920
-
Ring finger protein 145 (RNF145) is a ubiquitin ligase for sterol-induced degradation of HMG-CoA reductase.J Biol Chem. 2018 Mar 16;293(11):4047-4055. doi: 10.1074/jbc.RA117.001260. Epub 2018 Jan 26. J Biol Chem. 2018. PMID: 29374057 Free PMC article.
-
Srebf and Runx3 regulate hepatic HMG-CoA reductase and intracellular cholesterol under hypoxia.Cell Mol Life Sci. 2025 Jun 4;82(1):222. doi: 10.1007/s00018-025-05736-3. Cell Mol Life Sci. 2025. PMID: 40464811 Free PMC article.
-
Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase.Cell Res. 2008 Jun;18(6):609-21. doi: 10.1038/cr.2008.61. Cell Res. 2008. PMID: 18504457 Free PMC article. Review.
-
Underlying mechanisms for sterol-induced ubiquitination and ER-associated degradation of HMG CoA reductase.Semin Cell Dev Biol. 2018 Sep;81:121-128. doi: 10.1016/j.semcdb.2017.10.019. Epub 2017 Nov 7. Semin Cell Dev Biol. 2018. PMID: 29107682 Free PMC article. Review.
Cited by
-
Oxygen: viral friend or foe?Virol J. 2020 Jul 27;17(1):115. doi: 10.1186/s12985-020-01374-2. Virol J. 2020. PMID: 32718318 Free PMC article. Review.
-
The role of roxadustat in chronic kidney disease patients complicated with anemia.Korean J Intern Med. 2023 Mar;38(2):147-156. doi: 10.3904/kjim.2022.318. Epub 2023 Jan 2. Korean J Intern Med. 2023. PMID: 36588451 Free PMC article. Review.
-
A HIF independent oxygen-sensitive pathway for controlling cholesterol synthesis.Nat Commun. 2023 Aug 9;14(1):4816. doi: 10.1038/s41467-023-40541-1. Nat Commun. 2023. PMID: 37558666 Free PMC article.
-
Mechanisms and regulation of cholesterol homeostasis.Nat Rev Mol Cell Biol. 2020 Apr;21(4):225-245. doi: 10.1038/s41580-019-0190-7. Epub 2019 Dec 17. Nat Rev Mol Cell Biol. 2020. PMID: 31848472 Review.
-
A small-molecule inhibitor of hypoxia-inducible factor prolyl hydroxylase improves obesity, nephropathy and cardiomyopathy in obese ZSF1 rats.PLoS One. 2021 Aug 2;16(8):e0255022. doi: 10.1371/journal.pone.0255022. eCollection 2021. PLoS One. 2021. PMID: 34339435 Free PMC article.
References
-
- Goldstein J. L., DeBose-Boyd R. A., and Brown M. S. (2006) Protein sensors for membrane sterols. Cell 124, 35–46 - PubMed
-
- Sever N., Song B. L., Yabe D., Goldstein J. L., Brown M. S., and DeBose-Boyd R. A. (2003) Insig-dependent ubiquitination and degradation of mammalian 3-hydroxy-3-methylglutaryl-CoA reductase stimulated by sterols and geranylgeraniol. J. Biol. Chem. 278, 52479–52490 - PubMed
-
- Sever N., Yang T., Brown M. S., Goldstein J. L., and DeBose-Boyd R. A. (2003) Accelerated degradation of HMG CoA reductase mediated by binding of insig-1 to its sterol-sensing domain. Mol. Cell 11, 25–33 - PubMed
-
- Song B. L., Javitt N. B., and DeBose-Boyd R. A. (2005) Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 1, 179–189 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

