Using microsecond single-molecule FRET to determine the assembly pathways of T4 ssDNA binding protein onto model DNA replication forks
- PMID: 28416680
- PMCID: PMC5422767
- DOI: 10.1073/pnas.1619819114
Using microsecond single-molecule FRET to determine the assembly pathways of T4 ssDNA binding protein onto model DNA replication forks
Abstract
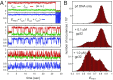
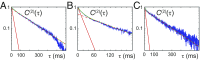
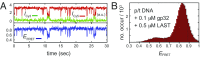
DNA replication is a core biological process that occurs in prokaryotic cells at high speeds (∼1 nucleotide residue added per millisecond) and with high fidelity (fewer than one misincorporation event per 107 nucleotide additions). The ssDNA binding protein [gene product 32 (gp32)] of the T4 bacteriophage is a central integrating component of the replication complex that must continuously bind to and unbind from transiently exposed template strands during DNA synthesis. We here report microsecond single-molecule FRET (smFRET) measurements on Cy3/Cy5-labeled primer-template (p/t) DNA constructs in the presence of gp32. These measurements probe the distance between Cy3/Cy5 fluorophores that label the ends of a short (15-nt) segment of ssDNA attached to a model p/t DNA construct and permit us to track the stochastic interconversion between various protein bound and unbound states. The length of the 15-nt ssDNA lattice is sufficient to accommodate up to two cooperatively bound gp32 proteins in either of two positions. We apply a unique multipoint time correlation function analysis to the microsecond-resolved smFRET data obtained to determine and compare the kinetics of various possible reaction pathways for the assembly of cooperatively bound gp32 protein onto ssDNA sequences located at the replication fork. The results of our analysis reveal the presence and translocation mechanisms of short-lived intermediate bound states that are likely to play a critical role in the assembly mechanisms of ssDNA binding proteins at replication forks and other ss duplex junctions.
Keywords: microsecond single-molecule FRET; multidimensional time correlation functions; ssDNA binding protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
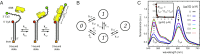
References
-
- Johnson A, O’Donnell M. Cellular DNA replicases: Components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. - PubMed
-
- Nossal NG. The bacteriphage T4 DNA replication fork. In: Karam JD, Kreuzer KN, Hall DH, editors. Molecular Biology of Bacteriphage T4. American Society for Microbiology; Washington, DC: 1994. pp. 43–54.
-
- Nossal NG, Makhov AM, Chastain PD, 2nd, Jones CE, Griffith JD. Architecture of the bacteriophage T4 replication complex revealed with nanoscale biopointers. J Biol Chem. 2007;282:1098–1108. - PubMed
-
- Alberts BM. Prokaryotic DNA replication mechanisms. Philos Trans R Soc Lond B Biol Sci. 1987;317:395–420. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

