The histone demethylase KDM3A regulates the transcriptional program of the androgen receptor in prostate cancer cells
- PMID: 28416760
- PMCID: PMC5444746
- DOI: 10.18632/oncotarget.15681
The histone demethylase KDM3A regulates the transcriptional program of the androgen receptor in prostate cancer cells
Abstract
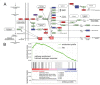
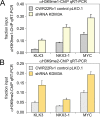
The lysine demethylase 3A (KDM3A, JMJD1A or JHDM2A) controls transcriptional networks in a variety of biological processes such as spermatogenesis, metabolism, stem cell activity, and tumor progression. We matched transcriptomic and ChIP-Seq profiles to decipher a genome-wide regulatory network of epigenetic control by KDM3A in prostate cancer cells. ChIP-Seq experiments monitoring histone 3 lysine 9 (H3K9) methylation marks show global histone demethylation effects of KDM3A. Combined assessment of histone demethylation events and gene expression changes presented major transcriptional activation suggesting that distinct oncogenic regulators may synergize with the epigenetic patterns by KDM3A. Pathway enrichment analysis of cells with KDM3A knockdown prioritized androgen signaling indicating that KDM3A plays a key role in regulating androgen receptor activity. Matched ChIP-Seq and knockdown experiments of KDM3A in combination with ChIP-Seq of the androgen receptor resulted in a gain of H3K9 methylation marks around androgen receptor binding sites of selected transcriptional targets in androgen signaling including positive regulation of KRT19, NKX3-1, KLK3, NDRG1, MAF, CREB3L4, MYC, INPP4B, PTK2B, MAPK1, MAP2K1, IGF1, E2F1, HSP90AA1, HIF1A, and ACSL3. The cancer systems biology analysis of KDM3A-dependent genes identifies an epigenetic and transcriptional network in androgen response, hypoxia, glycolysis, and lipid metabolism. Genome-wide ChIP-Seq data highlights specific gene targets and the ability of epigenetic master regulators to control oncogenic pathways and cancer progression.
Keywords: ChIP-Seq; cancer systems biology; epigenomics; oncogene; prostate cancer.
Conflict of interest statement
The authors declare that there is no competing interest as part of the submission process of this manuscript.
Figures


Similar articles
-
Global histone modification profiling reveals the epigenomic dynamics during malignant transformation in a four-stage breast cancer model.Clin Epigenetics. 2016 Mar 31;8:34. doi: 10.1186/s13148-016-0201-x. eCollection 2016. Clin Epigenetics. 2016. PMID: 27034728 Free PMC article.
-
Systematic knockdown of epigenetic enzymes identifies a novel histone demethylase PHF8 overexpressed in prostate cancer with an impact on cell proliferation, migration and invasion.Oncogene. 2012 Jul 19;31(29):3444-56. doi: 10.1038/onc.2011.512. Epub 2011 Nov 28. Oncogene. 2012. PMID: 22120715
-
Regulation of c-Myc expression by the histone demethylase JMJD1A is essential for prostate cancer cell growth and survival.Oncogene. 2016 May 12;35(19):2441-52. doi: 10.1038/onc.2015.309. Epub 2015 Aug 17. Oncogene. 2016. PMID: 26279298 Free PMC article.
-
Sequencing the transcriptional network of androgen receptor in prostate cancer.Cancer Lett. 2013 Nov 1;340(2):254-60. doi: 10.1016/j.canlet.2012.11.009. Epub 2012 Nov 27. Cancer Lett. 2013. PMID: 23196061 Review.
-
Role of ChIP-seq in the discovery of transcription factor binding sites, differential gene regulation mechanism, epigenetic marks and beyond.Cell Cycle. 2014;13(18):2847-52. doi: 10.4161/15384101.2014.949201. Cell Cycle. 2014. PMID: 25486472 Free PMC article. Review.
Cited by
-
Histone demethylase JMJD1A promotes alternative splicing of AR variant 7 (AR-V7) in prostate cancer cells.Proc Natl Acad Sci U S A. 2018 May 15;115(20):E4584-E4593. doi: 10.1073/pnas.1802415115. Epub 2018 Apr 30. Proc Natl Acad Sci U S A. 2018. PMID: 29712835 Free PMC article.
-
AR-negative prostate cancer is vulnerable to loss of JMJD1C demethylase.Proc Natl Acad Sci U S A. 2021 Sep 7;118(36):e2026324118. doi: 10.1073/pnas.2026324118. Proc Natl Acad Sci U S A. 2021. PMID: 34475205 Free PMC article.
-
DOT1L: a key target in normal chromatin remodelling and in mixed-lineage leukaemia treatment.Epigenetics. 2020 May;15(5):439-453. doi: 10.1080/15592294.2019.1699991. Epub 2019 Dec 28. Epigenetics. 2020. PMID: 31790636 Free PMC article. Review.
-
KDM3A-mediated SP1 activates PFKFB4 transcription to promote aerobic glycolysis in osteosarcoma and augment tumor development.BMC Cancer. 2022 May 19;22(1):562. doi: 10.1186/s12885-022-09636-8. BMC Cancer. 2022. PMID: 35590288 Free PMC article.
-
Advances in Histone Demethylase KDM3A as a Cancer Therapeutic Target.Cancers (Basel). 2020 Apr 28;12(5):1098. doi: 10.3390/cancers12051098. Cancers (Basel). 2020. PMID: 32354028 Free PMC article. Review.
References
-
- Gonda TJ, Ramsay RG. Directly targeting transcriptional dysregulation in cancer. Nat Rev Cancer. 2015;15:686–94. - PubMed
-
- Kooistra SM, Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol. 2012;13:297–311. - PubMed
-
- Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–95. - PubMed
-
- Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–23. - PubMed
-
- Kuroki S, Matoba S, Akiyoshi M, Matsumura Y, Miyachi H, Mise N, Abe K, Ogura A, Wilhelm D, Koopman P, Nozaki M, Kanai Y, Shinkai Y, et al. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science. 2013;341:1106–09. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

