Use of Graph Database for the Integration of Heterogeneous Biological Data
- PMID: 28416946
- PMCID: PMC5389944
- DOI: 10.5808/GI.2017.15.1.19
Use of Graph Database for the Integration of Heterogeneous Biological Data
Abstract
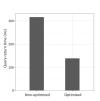
Understanding complex relationships among heterogeneous biological data is one of the fundamental goals in biology. In most cases, diverse biological data are stored in relational databases, such as MySQL and Oracle, which store data in multiple tables and then infer relationships by multiple-join statements. Recently, a new type of database, called the graph-based database, was developed to natively represent various kinds of complex relationships, and it is widely used among computer science communities and IT industries. Here, we demonstrate the feasibility of using a graph-based database for complex biological relationships by comparing the performance between MySQL and Neo4j, one of the most widely used graph databases. We collected various biological data (protein-protein interaction, drug-target, gene-disease, etc.) from several existing sources, removed duplicate and redundant data, and finally constructed a graph database containing 114,550 nodes and 82,674,321 relationships. When we tested the query execution performance of MySQL versus Neo4j, we found that Neo4j outperformed MySQL in all cases. While Neo4j exhibited a very fast response for various queries, MySQL exhibited latent or unfinished responses for complex queries with multiple-join statements. These results show that using graph-based databases, such as Neo4j, is an efficient way to store complex biological relationships. Moreover, querying a graph database in diverse ways has the potential to reveal novel relationships among heterogeneous biological data.
Keywords: Neo4j; biological network; data mining; graph database; heterogeneous biological data; query performance.
Figures







References
-
- Hartwell LH, Hopfield JJ, Leibler S, Murray AW. From molecular to modular cell biology. Nature. 1999;402(6761) Suppl:C47–C52. - PubMed
-
- Kitano H. Computational systems biology. Nature. 2002;420:206–210. - PubMed
-
- Koonin EV, Wolf YI, Karev GP. The structure of the protein universe and genome evolution. Nature. 2002;420:218–223. - PubMed
-
- Alon U. Biological networks: the tinkerer as an engineer. Science. 2003;301:1866–1867. - PubMed
-
- Bray D. Molecular networks: the top-down view. Science. 2003;301:1864–1865. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
