DNA methylation profiling in peripheral lung tissues of smokers and patients with COPD
- PMID: 28416970
- PMCID: PMC5391602
- DOI: 10.1186/s13148-017-0335-5
DNA methylation profiling in peripheral lung tissues of smokers and patients with COPD
Abstract
Background: Epigenetics changes have been shown to be affected by cigarette smoking. Cigarette smoke (CS)-mediated DNA methylation can potentially affect several cellular and pathophysiological processes, acute exacerbations, and comorbidity in the lungs of patients with chronic obstructive pulmonary disease (COPD). We sought to determine whether genome-wide lung DNA methylation profiles of smokers and patients with COPD were significantly different from non-smokers. We isolated DNA from parenchymal lung tissues of patients including eight lifelong non-smokers, eight current smokers, and eight patients with COPD and analyzed the samples using Illumina's Infinium HumanMethylation450 BeadChip.
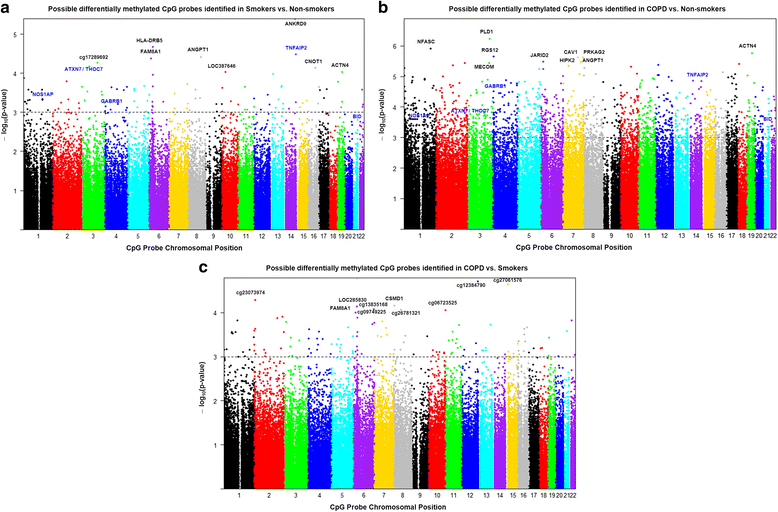
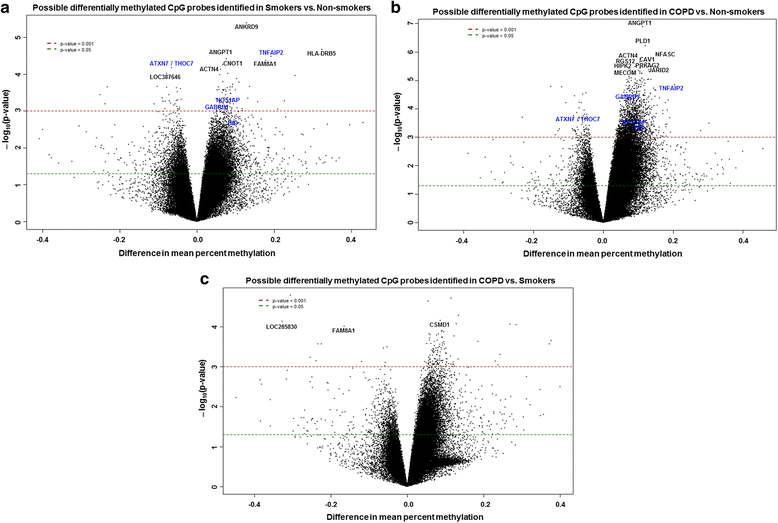
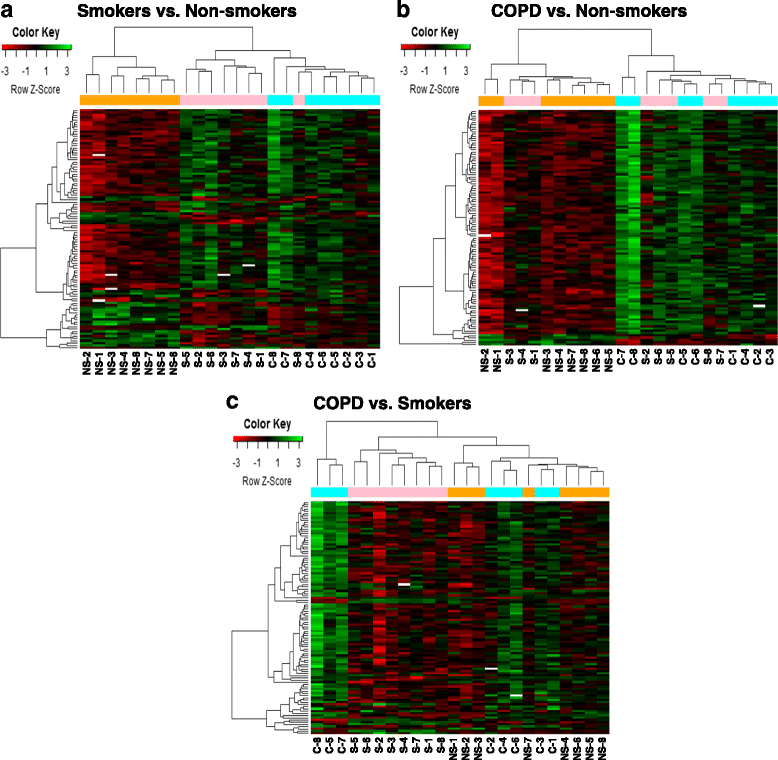
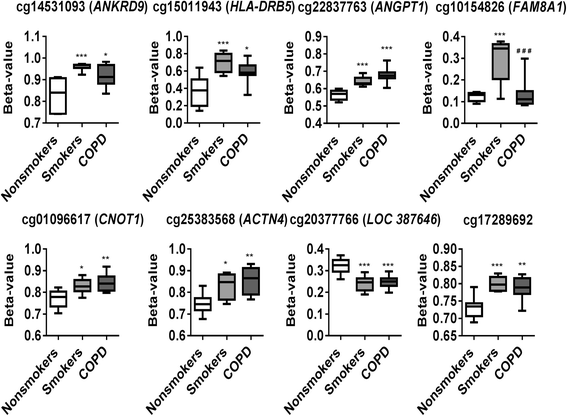
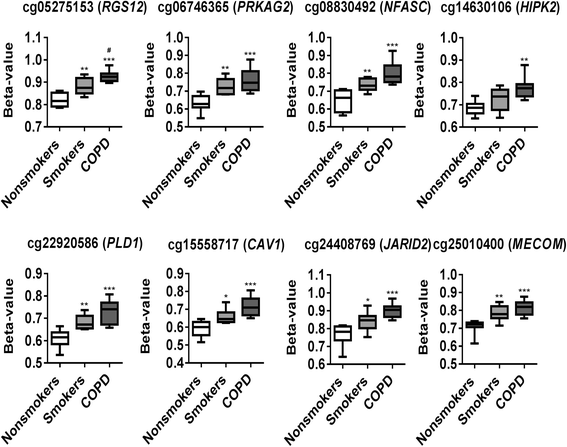
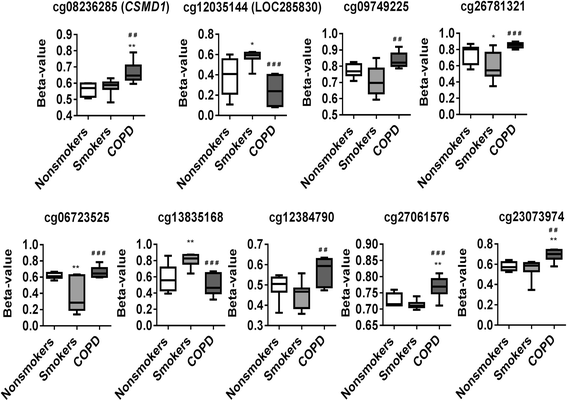
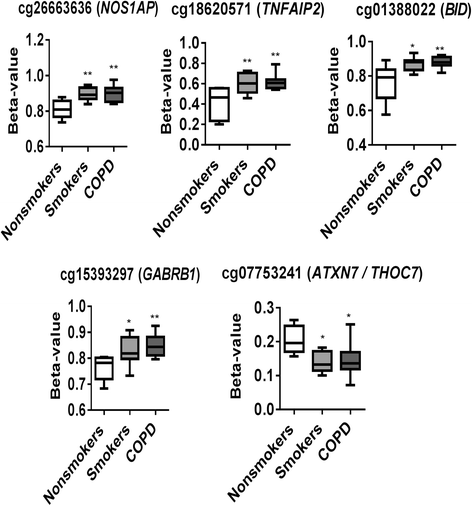
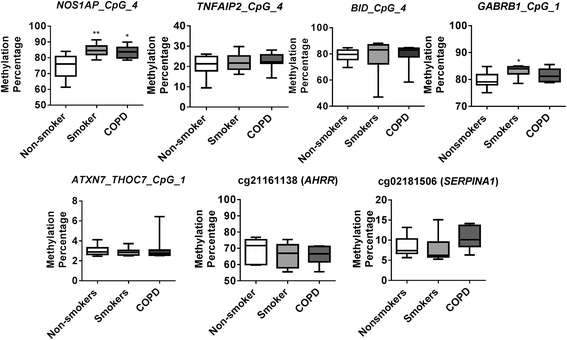
Results: Our data revealed that the differentially methylated genes were related to top canonical pathways (e.g., G beta gamma signaling, mechanisms of cancer, and nNOS signaling in neurons), disease and disorders (organismal injury and abnormalities, cancer, and respiratory disease), and molecular and cellular functions (cell death and survival, cellular assembly and organization, cellular function and maintenance) in patients with COPD. The genome-wide DNA methylation analysis identified suggestive genes, such as NOS1AP, TNFAIP2, BID, GABRB1, ATXN7, and THOC7 with DNA methylation changes in COPD lung tissues that were further validated by pyrosequencing. Pyrosequencing validation confirmed hyper-methylation in smokers and patients with COPD as compared to non-smokers. However, we did not detect significant differences in DNA methylation for TNFAIP2, ATXN7, and THOC7 genes in smokers and COPD groups despite the changes observed in the genome-wide analysis.
Conclusions: Our study suggests that DNA methylation in suggestive genes, such as NOS1AP, BID, and GABRB1 may be used as epigenetic signatures in smokers and patients with COPD if the same is validated in a larger cohort. Future studies are required to correlate DNA methylation status with transcriptomics of selective genes identified in this study and elucidate their role and involvement in the progression of COPD and its exacerbations.
Keywords: COPD; DNA methylation; Epigenetics; Lung; Pyrosequencing; Smokers.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

