Radiogenomic Analysis of Oncological Data: A Technical Survey
- PMID: 28417933
- PMCID: PMC5412389
- DOI: 10.3390/ijms18040805
Radiogenomic Analysis of Oncological Data: A Technical Survey
Abstract
In the last few years, biomedical research has been boosted by the technological development of analytical instrumentation generating a large volume of data. Such information has increased in complexity from basic (i.e., blood samples) to extensive sets encompassing many aspects of a subject phenotype, and now rapidly extending into genetic and, more recently, radiomic information. Radiogenomics integrates both aspects, investigating the relationship between imaging features and gene expression. From a methodological point of view, radiogenomics takes advantage of non-conventional data analysis techniques that reveal meaningful information for decision-support in cancer diagnosis and treatment. This survey is aimed to review the state-of-the-art techniques employed in radiomics and genomics with special focus on analysis methods based on molecular and multimodal probes. The impact of single and combined techniques will be discussed in light of their suitability in correlation and predictive studies of specific oncologic diseases.
Keywords: MR; NGS technologies; cancer; correlation matrix; data mining; microarray; molecular imaging; radiogenomics; texture analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
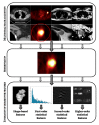
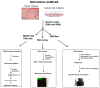
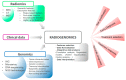
References
-
- Elsheikh T.M., Asa S.L., Chan J.K.C., DeLellis R.A., Heffess C.S., LiVolsi V.A., Wenig B.M. Interobserver and intraobserver variation among experts in the diagnosis of thyroid follicular lesions with borderline nuclear features of papillary carcinoma. Am. J. Clin. Pathol. 2008;130:736–744. doi: 10.1309/AJCPKP2QUVN4RCCP. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

