Accurate and reproducible invasive breast cancer detection in whole-slide images: A Deep Learning approach for quantifying tumor extent
- PMID: 28418027
- PMCID: PMC5394452
- DOI: 10.1038/srep46450
Accurate and reproducible invasive breast cancer detection in whole-slide images: A Deep Learning approach for quantifying tumor extent
Abstract
With the increasing ability to routinely and rapidly digitize whole slide images with slide scanners, there has been interest in developing computerized image analysis algorithms for automated detection of disease extent from digital pathology images. The manual identification of presence and extent of breast cancer by a pathologist is critical for patient management for tumor staging and assessing treatment response. However, this process is tedious and subject to inter- and intra-reader variability. For computerized methods to be useful as decision support tools, they need to be resilient to data acquired from different sources, different staining and cutting protocols and different scanners. The objective of this study was to evaluate the accuracy and robustness of a deep learning-based method to automatically identify the extent of invasive tumor on digitized images. Here, we present a new method that employs a convolutional neural network for detecting presence of invasive tumor on whole slide images. Our approach involves training the classifier on nearly 400 exemplars from multiple different sites, and scanners, and then independently validating on almost 200 cases from The Cancer Genome Atlas. Our approach yielded a Dice coefficient of 75.86%, a positive predictive value of 71.62% and a negative predictive value of 96.77% in terms of pixel-by-pixel evaluation compared to manually annotated regions of invasive ductal carcinoma.
Conflict of interest statement
Drs Madabhushi, Feldman, Ganesan, and Tomaszewski are scientific consultants for the digital pathology company Inspirata Inc. Drs Madabhushi, Feldman, Ganesan, and Tomaszewski also serve on the scientific advisory board for the digital pathology company Inspirata Inc. Dr. Madabhushi also has an equity stake in Inspirata Inc. and Elucid Bioimaging Inc.
Figures



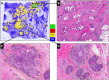



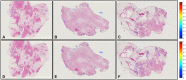



References
-
- Genestie C. et al.. Comparison of the prognostic value of Scarff-Bloom-Richardson and nottingham histological grades in a series of 825 cases of breast cancer: major importance of the mitotic count as a component of both grading systems. Anticancer Research 18, 571–576 (1998). - PubMed
-
- Elston C. W. & Ellis I. O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19, 403–410 (1991). - PubMed
-
- Frierson H. F. et al.. Interobserver reproducibility of the Nottingham modification of the Bloom and Richardson histologic grading scheme for infiltrating ductal carcinoma. American journal of clinical pathology 103, 195–8 (1995). - PubMed
-
- Gomes D. S., Porto S. S., Balabram D. & Gobbi H. Inter-observer variability between general pathologists and a specialist in breast pathology in the diagnosis of lobular neoplasia, columnar cell lesions, atypical ductal hyperplasia and ductal carcinoma in situ of the breast. Diagnostic pathology 9, 121 (2014). - PMC - PubMed
-
- Longacre T. A. et al.. Interobserver agreement and reproducibility in classification of invasive breast carcinoma: an NCI breast cancer family registry study. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc 19, 195–207 (2006). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

