Erroneous energy-generating cycles in published genome scale metabolic networks: Identification and removal
- PMID: 28419089
- PMCID: PMC5413070
- DOI: 10.1371/journal.pcbi.1005494
Erroneous energy-generating cycles in published genome scale metabolic networks: Identification and removal
Abstract
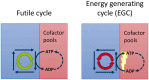
Energy metabolism is central to cellular biology. Thus, genome-scale models of heterotrophic unicellular species must account appropriately for the utilization of external nutrients to synthesize energy metabolites such as ATP. However, metabolic models designed for flux-balance analysis (FBA) may contain thermodynamically impossible energy-generating cycles: without nutrient consumption, these models are still capable of charging energy metabolites (such as ADP→ATP or NADP+→NADPH). Here, we show that energy-generating cycles occur in over 85% of metabolic models without extensive manual curation, such as those contained in the ModelSEED and MetaNetX databases; in contrast, such cycles are rare in the manually curated models of the BiGG database. Energy generating cycles may represent model errors, e.g., erroneous assumptions on reaction reversibilities. Alternatively, part of the cycle may be thermodynamically feasible in one environment, while the remainder is thermodynamically feasible in another environment; as standard FBA does not account for thermodynamics, combining these into an FBA model allows erroneous energy generation. The presence of energy-generating cycles typically inflates maximal biomass production rates by 25%, and may lead to biases in evolutionary simulations. We present efficient computational methods (i) to identify energy generating cycles, using FBA, and (ii) to identify minimal sets of model changes that eliminate them, using a variant of the GlobalFit algorithm.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

