Enhancing faba bean (Vicia faba L.) genome resources
- PMID: 28419381
- PMCID: PMC5429004
- DOI: 10.1093/jxb/erx117
Enhancing faba bean (Vicia faba L.) genome resources
Abstract
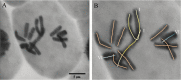
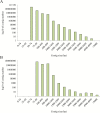
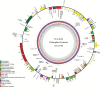
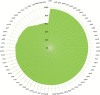
Grain legume improvement is currently impeded by a lack of genomic resources. The paucity of genome information for faba bean can be attributed to the intrinsic difficulties of assembling/annotating its giant (~13 Gb) genome. In order to address this challenge, RNA-sequencing analysis was performed on faba bean (cv. Wizard) leaves. Read alignment to the faba bean reference transcriptome identified 16 300 high quality unigenes. In addition, Illumina paired-end sequencing was used to establish a baseline for genomic information assembly. Genomic reads were assembled de novo into contigs with a size range of 50-5000 bp. Over 85% of sequences did not align to known genes, of which ~10% could be aligned to known repetitive genetic elements. Over 26 000 of the reference transcriptome unigenes could be aligned to DNA-sequencing (DNA-seq) reads with high confidence. Moreover, this comparison identified 56 668 potential splice points in all identified unigenes. Sequence length data were extended at 461 putative loci through alignment of DNA-seq contigs to full-length, publicly available linkage marker sequences. Reads also yielded coverages of 3466× and 650× for the chloroplast and mitochondrial genomes, respectively. Inter- and intraspecies organelle genome comparisons established core legume organelle gene sets, and revealed polymorphic regions of faba bean organelle genomes.
Keywords: Illumina sequencing; RNA-seq analysis.; legumes; mitochondrial genome; plastome; protein security.
© The Author 2017. Published by Oxford University Press on behalf of the Society for Experimental Biology.
Figures


References
-
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Retrieved on 4....
-
- Barton L, Thamo T, Engelbrecht D, Biswas WK. 2014. Does growing grain legumes or applying lime cost effectively lower greenhouse gas emissions from wheat production in a semi-arid climate? Journal of Cleaner Production 83, 194–203.
-
- Benedito VA, Torres-Jerez I, Murray JD, et al. 2008. A gene expression atlas of the model legume Medicago truncatula. The Plant Journal 55, 504–513. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

