Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden
- PMID: 28420421
- PMCID: PMC5395719
- DOI: 10.1186/s13073-017-0424-2
Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden
Abstract
Background: High tumor mutational burden (TMB) is an emerging biomarker of sensitivity to immune checkpoint inhibitors and has been shown to be more significantly associated with response to PD-1 and PD-L1 blockade immunotherapy than PD-1 or PD-L1 expression, as measured by immunohistochemistry (IHC). The distribution of TMB and the subset of patients with high TMB has not been well characterized in the majority of cancer types.
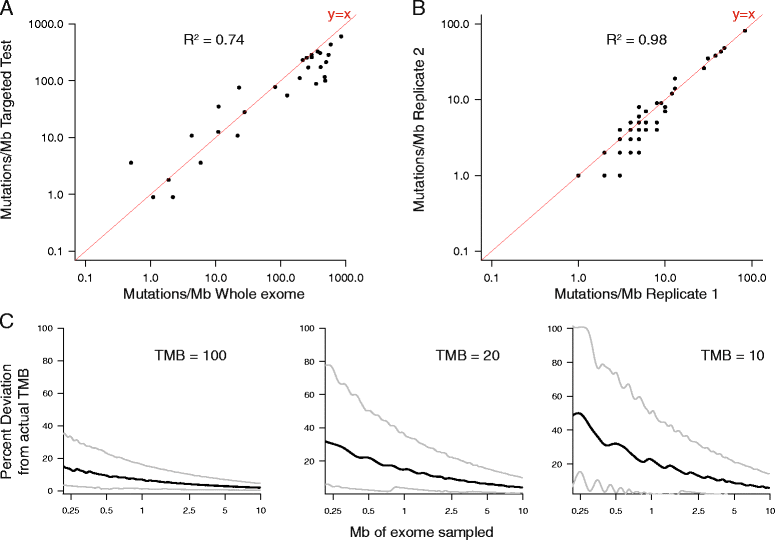
Methods: In this study, we compare TMB measured by a targeted comprehensive genomic profiling (CGP) assay to TMB measured by exome sequencing and simulate the expected variance in TMB when sequencing less than the whole exome. We then describe the distribution of TMB across a diverse cohort of 100,000 cancer cases and test for association between somatic alterations and TMB in over 100 tumor types.
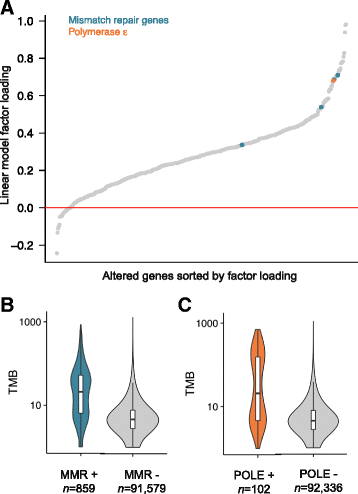
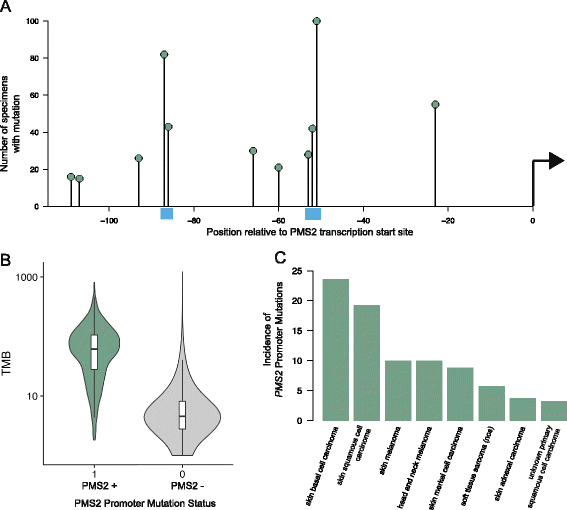
Results: We demonstrate that measurements of TMB from comprehensive genomic profiling are strongly reflective of measurements from whole exome sequencing and model that below 0.5 Mb the variance in measurement increases significantly. We find that a subset of patients exhibits high TMB across almost all types of cancer, including many rare tumor types, and characterize the relationship between high TMB and microsatellite instability status. We find that TMB increases significantly with age, showing a 2.4-fold difference between age 10 and age 90 years. Finally, we investigate the molecular basis of TMB and identify genes and mutations associated with TMB level. We identify a cluster of somatic mutations in the promoter of the gene PMS2, which occur in 10% of skin cancers and are highly associated with increased TMB.
Conclusions: These results show that a CGP assay targeting ~1.1 Mb of coding genome can accurately assess TMB compared with sequencing the whole exome. Using this method, we find that many disease types have a substantial portion of patients with high TMB who might benefit from immunotherapy. Finally, we identify novel, recurrent promoter mutations in PMS2, which may be another example of regulatory mutations contributing to tumorigenesis.
Keywords: Cancer genomics; Mismatch repair; PMS2; Tumor mutational burden.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

