Expression of long non-coding RNAs in autoimmunity and linkage to enhancer function and autoimmune disease risk genetic variants
- PMID: 28420548
- PMCID: PMC5508524
- DOI: 10.1016/j.jaut.2017.03.014
Expression of long non-coding RNAs in autoimmunity and linkage to enhancer function and autoimmune disease risk genetic variants
Abstract
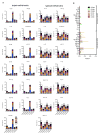
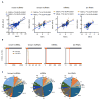
Genome-wide association studies have identified numerous genetic variants conferring autoimmune disease risk. Most of these genetic variants lie outside protein-coding genes hampering mechanistic explorations. Numerous mRNAs are also differentially expressed in autoimmune disease but their regulation is also unclear. The majority of the human genome is transcribed yet its biologic significance is incompletely understood. We performed whole genome RNA-sequencing [RNA-seq] to categorize expression of mRNAs, known and novel long non-coding RNAs [lncRNAs] in leukocytes from subjects with autoimmune disease and identified annotated and novel lncRNAs differentially expressed across multiple disorders. We found that loci transcribing novel lncRNAs were not randomly distributed across the genome but co-localized with leukocyte transcriptional enhancers, especially super-enhancers, and near genetic variants associated with autoimmune disease risk. We propose that alterations in enhancer function, including lncRNA expression, produced by genetics and environment, change cellular phenotypes contributing to disease risk and pathogenesis and represent attractive therapeutic targets.
Copyright © 2017 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

