Structure and characterization of a class 3B proline utilization A: Ligand-induced dimerization and importance of the C-terminal domain for catalysis
- PMID: 28420730
- PMCID: PMC5465489
- DOI: 10.1074/jbc.M117.786855
Structure and characterization of a class 3B proline utilization A: Ligand-induced dimerization and importance of the C-terminal domain for catalysis
Abstract
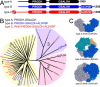
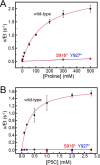
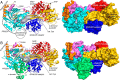
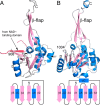
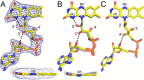
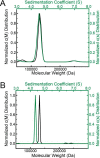
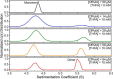
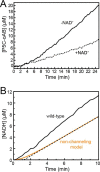
The bifunctional flavoenzyme proline utilization A (PutA) catalyzes the two-step oxidation of proline to glutamate using separate proline dehydrogenase (PRODH) and l-glutamate-γ-semialdehyde dehydrogenase active sites. Because PutAs catalyze sequential reactions, they are good systems for studying how metabolic enzymes communicate via substrate channeling. Although mechanistically similar, PutAs vary widely in domain architecture, oligomeric state, and quaternary structure, and these variations represent different structural solutions to the problem of sequestering a reactive metabolite. Here, we studied PutA from Corynebacterium freiburgense (CfPutA), which belongs to the uncharacterized 3B class of PutAs. A 2.7 Å resolution crystal structure showed the canonical arrangement of PRODH, l-glutamate-γ-semialdehyde dehydrogenase, and C-terminal domains, including an extended interdomain tunnel associated with substrate channeling. The structure unexpectedly revealed a novel open conformation of the PRODH active site, which is interpreted to represent the non-activated conformation, an elusive form of PutA that exhibits suboptimal channeling. Nevertheless, CfPutA exhibited normal substrate-channeling activity, indicating that it isomerizes into the active state under assay conditions. Sedimentation-velocity experiments provided insight into the isomerization process, showing that CfPutA dimerizes in the presence of a proline analog and NAD+ These results are consistent with the morpheein model of enzyme hysteresis, in which substrate binding induces conformational changes that promote assembly of a high-activity oligomer. Finally, we used domain deletion analysis to investigate the function of the C-terminal domain. Although this domain contains neither catalytic residues nor substrate sites, its removal impaired both catalytic activities, suggesting that it may be essential for active-site integrity.
Keywords: X-ray crystallography; analytical ultracentrifugation; bifunctional enzyme; dehydrogenase; enzyme kinetics; flavoprotein; oligomerization; substrate channeling.
© 2017 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

References
-
- Tanner J. J., and Becker D. F. (2013) PutA and proline metabolism. in Handbook of Flavoproteins, Vol. 1 (Hille R., Miller S. M., and Palfey B. A., eds) pp. 31–56, De Gruyter, Berlin
-
- Srivastava D., Schuermann J. P., White T. A., Krishnan N., Sanyal N., Hura G. L., Tan A., Henzl M. T., Becker D. F., and Tanner J. J. (2010) Crystal structure of the bifunctional proline utilization A flavoenzyme from Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. U.S.A. 107, 2878–2883 - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

