Hierarchical assembly of centriole subdistal appendages via centrosome binding proteins CCDC120 and CCDC68
- PMID: 28422092
- PMCID: PMC5399293
- DOI: 10.1038/ncomms15057
Hierarchical assembly of centriole subdistal appendages via centrosome binding proteins CCDC120 and CCDC68
Abstract
In animal cells, the centrosome is the main microtubule-organizing centre where microtubules are nucleated and anchored. The centriole subdistal appendages (SDAs) are the key structures that anchor microtubules in interphase cells, but the composition and assembly mechanisms of SDAs are not well understood. Here, we reveal that centrosome-binding proteins, coiled-coil domain containing (CCDC) 120 and CCDC68 are two novel SDA components required for hierarchical SDA assembly in human cells. CCDC120 is anchored to SDAs by ODF2 and recruits CEP170 and Ninein to the centrosome through different coiled-coil domains at its N terminus. CCDC68 is a CEP170-interacting protein that competes with CCDC120 in recruiting CEP170 to SDAs. Furthermore, CCDC120 and CCDC68 are required for centrosome microtubule anchoring. Our findings elucidate the molecular basis for centriole SDA hierarchical assembly and microtubule anchoring in human interphase cells.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

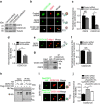
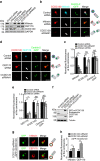


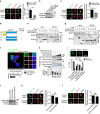
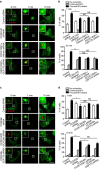

References
-
- Doxsey S. Re-evaluating centrosome function. Nat. Rev. Mol. Cell Biol. 2, 688–698 (2001). - PubMed
-
- Bettencourt-Dias M. & Glover D. M. Centrosome biogenesis and function: centrosomics brings new understanding. Nat. Rev. Mol. Cell Biol. 8, 451–463 (2007). - PubMed
-
- Nigg E. A. & Raff J. W. Centrioles, centrosomes, and cilia in health and disease. Cell 139, 663–678 (2009). - PubMed
-
- Piel M., Nordberg J., Euteneuer U. & Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science 291, 1550–1553 (2001). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

