Genetic structure and signatures of selection in grey reef sharks (Carcharhinus amblyrhynchos)
- PMID: 28422134
- PMCID: PMC5555095
- DOI: 10.1038/hdy.2017.21
Genetic structure and signatures of selection in grey reef sharks (Carcharhinus amblyrhynchos)
Abstract
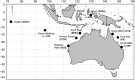
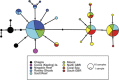
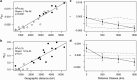
With overfishing reducing the abundance of marine predators in multiple marine ecosystems, knowledge of genetic structure and local adaptation may provide valuable information to assist sustainable management. Despite recent technological advances, most studies on sharks have used small sets of neutral markers to describe their genetic structure. We used 5517 nuclear single-nucleotide polymorphisms (SNPs) and a mitochondrial DNA (mtDNA) gene to characterize patterns of genetic structure and detect signatures of selection in grey reef sharks (Carcharhinus amblyrhynchos). Using samples from Australia, Indonesia and oceanic reefs in the Indian Ocean, we established that large oceanic distances represent barriers to gene flow, whereas genetic differentiation on continental shelves follows an isolation by distance model. In Australia and Indonesia differentiation at nuclear SNPs was weak, with coral reefs acting as stepping stones maintaining connectivity across large distances. Differentiation of mtDNA was stronger, and more pronounced in females, suggesting sex-biased dispersal. Four independent tests identified a set of loci putatively under selection, indicating that grey reef sharks in eastern Australia are likely under different selective pressures to those in western Australia and Indonesia. Genetic distances averaged across all loci were uncorrelated with genetic distances calculated from outlier loci, supporting the conclusion that different processes underpin genetic divergence in these two data sets. This pattern of heterogeneous genomic differentiation, suggestive of local adaptation, has implications for the conservation of grey reef sharks; furthermore, it highlights that marine species showing little genetic differentiation at neutral loci may exhibit patterns of cryptic genetic structure driven by local selection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Contrasting movements and connectivity of reef-associated sharks using acoustic telemetry: implications for management.Ecol Appl. 2015 Dec;25(8):2101-18. doi: 10.1890/14-2293.1. Ecol Appl. 2015. PMID: 26910942
-
Blacktip reef sharks, Carcharhinus melanopterus, have high genetic structure and varying demographic histories in their Indo-Pacific range.Mol Ecol. 2014 Nov;23(21):5193-207. doi: 10.1111/mec.12936. Epub 2014 Oct 13. Mol Ecol. 2014. PMID: 25251515
-
Connectivity in grey reef sharks (Carcharhinus amblyrhynchos) determined using empirical and simulated genetic data.Sci Rep. 2015 Aug 28;5:13229. doi: 10.1038/srep13229. Sci Rep. 2015. PMID: 26314287 Free PMC article.
-
Larval dispersal and movement patterns of coral reef fishes, and implications for marine reserve network design.Biol Rev Camb Philos Soc. 2015 Nov;90(4):1215-47. doi: 10.1111/brv.12155. Epub 2014 Nov 25. Biol Rev Camb Philos Soc. 2015. PMID: 25423947 Review.
-
Phylogeography of sharks and rays: a global review based on life history traits and biogeographic partitions.PeerJ. 2023 Jun 1;11:e15396. doi: 10.7717/peerj.15396. eCollection 2023. PeerJ. 2023. PMID: 37283899 Free PMC article. Review.
Cited by
-
Contrasting global, regional and local patterns of genetic structure in gray reef shark populations from the Indo-Pacific region.Sci Rep. 2019 Nov 1;9(1):15816. doi: 10.1038/s41598-019-52221-6. Sci Rep. 2019. PMID: 31676818 Free PMC article.
-
Like a rolling stone: Colonization and migration dynamics of the gray reef shark (Carcharhinus amblyrhynchos).Ecol Evol. 2023 Jan 10;13(1):e9746. doi: 10.1002/ece3.9746. eCollection 2023 Jan. Ecol Evol. 2023. PMID: 36644707 Free PMC article.
-
Seascape Genomics of the Smooth Hammerhead Shark Sphyrna zygaena Reveals Regional Adaptive Clinal Variation.Ecol Evol. 2024 Dec 12;14(12):e70644. doi: 10.1002/ece3.70644. eCollection 2024 Dec. Ecol Evol. 2024. PMID: 39669504 Free PMC article.
-
Fine-scale temperature-associated genetic structure between inshore and offshore populations of sea scallop (Placopecten magellanicus).Heredity (Edinb). 2019 Jan;122(1):69-80. doi: 10.1038/s41437-018-0087-9. Epub 2018 May 17. Heredity (Edinb). 2019. PMID: 29773897 Free PMC article.
-
Genomic evidence indicates small island-resident populations and sex-biased behaviors of Hawaiian reef Manta Rays.BMC Ecol Evol. 2023 Jul 8;23(1):31. doi: 10.1186/s12862-023-02130-0. BMC Ecol Evol. 2023. PMID: 37422622 Free PMC article.
References
-
- Ahonen H, Lowther AD, Harcourt RG, Goldsworthy SD, Charrier I, Stow AJ. (2016). The limits of dispersal: fine scale spatial genetic structure in Australian sea lions. Front Mar Sci 3: 65.
-
- Allendorf FW, Hohenlohe PA, Luikart G. (2010). Genomics and the future of conservation genetics. Nat Rev Genet 11: 697–709. - PubMed
-
- Almany G, Connolly S, Heath D, Hogan J, Jones G, McCook L et al. (2009). Connectivity, biodiversity conservation and the design of marine reserve networks for coral reefs. Coral Reefs 28: 339–351.
-
- Arèvalo E, Davis SK, Sites JW. (1994). Mitochondrial DNA sequence divergence and phylogenetic relationships among eight chromosome races of the Sceloporus grammicus complex (Phrynosomatidae) in central Mexico. Syst Biol 43: 387–418.
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources

