Exome Analysis of Rare and Common Variants within the NOD Signaling Pathway
- PMID: 28422189
- PMCID: PMC5396125
- DOI: 10.1038/srep46454
Exome Analysis of Rare and Common Variants within the NOD Signaling Pathway
Abstract
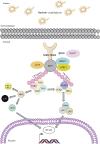
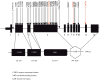
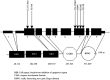
Pediatric inflammatory bowel disease (pIBD) is a chronic heterogeneous disorder. This study looks at the burden of common and rare coding mutations within 41 genes comprising the NOD signaling pathway in pIBD patients. 136 pIBD and 106 control samples underwent whole-exome sequencing. We compared the burden of common, rare and private mutation between these two groups using the SKAT-O test. An independent replication cohort of 33 cases and 111 controls was used to validate significant findings. We observed variation in 40 of 41 genes comprising the NOD signaling pathway. Four genes were significantly associated with disease in the discovery cohort (BIRC2 p = 0.004, NFKB1 p = 0.005, NOD2 p = 0.029 and SUGT1 p = 0.047). Statistical significance was replicated for BIRC2 (p = 0.041) and NOD2 (p = 0.045) in an independent validation cohort. A gene based test on the combined discovery and replication cohort confirmed association for BIRC2 (p = 0.030). We successfully applied burden of mutation testing that jointly assesses common and rare variants, identifying two previously implicated genes (NFKB1 and NOD2) and confirmed a possible role in disease risk in a previously unreported gene (BIRC2). The identification of this novel gene provides a wider role for the inhibitor of apoptosis gene family in IBD pathogenesis.
Conflict of interest statement
We confirm that this manuscript has not been published elsewhere, and is not under consideration by another journal. All the authors have read and approved the manuscript, and there is no ethical problem or conflict of interest with regard to this manuscript.
The authors declare no competing financial interests.
Figures
References
-
- Ruel J., Ruane D., Mehandru S., Gower-Rousseau C. & Colombel J.-F. IBD across the age spectrum-is it the same disease? Nat. Rev. Gastroenterol. Hepatol. 11, 88–98 (2014). - PubMed
-
- Hugot J. P. et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411, 599–603 (2001). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

