Interrelationships of VEL1 and ENV1 in light response and development in Trichoderma reesei
- PMID: 28423024
- PMCID: PMC5397039
- DOI: 10.1371/journal.pone.0175946
Interrelationships of VEL1 and ENV1 in light response and development in Trichoderma reesei
Abstract
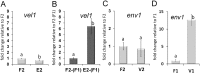
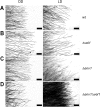
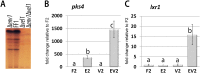
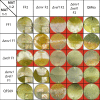
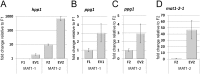
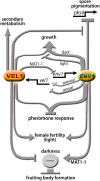
Sexual development is regulated by a complex regulatory mechanism in fungi. For Trichoderma reesei, the light response pathway was shown to impact sexual development, particularly through the photoreceptor ENVOY. Moreover, T. reesei communicates chemically with a potential mating partner in its vicinity, a response which is mediated by the velvet family protein VEL1 and its impact on secondary metabolism. We therefore studied the regulatory interactions of ENV1 and VEL1 with a focus on sexual development. Although individual mutants in both genes are female sterile under standard crossing conditions (light-dark cycles), an altered light regime enabled sexual development, which we found to be due to conditional female sterility of Δenv1, but not Δvel1. Phenotypes of growth and asexual sporulation as well as regulation of the peptide pheromone precursors of double mutants suggested that ENV1 and VEL1 balance positive and negative regulators of these functions. Additionally, VEL1 contributed to the strong deregulation of the pheromone system observed in env1 mutants. Female sterility of Δvel1 was rescued by deletion of env1 in darkness in MAT1-1, indicating a block of sexual development by ENV1 in darkness that is balanced by VEL1 in the wild-type. We conclude that ENV1 and VEL1 exert complementing functions in development of T. reesei. Our results further showed that the different developmental phenotypes of vel1/veA mutants in T. reesei and Aspergillus nidulans are not due to the presence or function of ENV1 in the VELVET regulatory pathway in T. reesei.
Conflict of interest statement
Figures



Similar articles
-
Light, stress, sex and carbon - The photoreceptor ENVOY as a central checkpoint in the physiology of Trichoderma reesei.Fungal Biol. 2018 Jun;122(6):479-486. doi: 10.1016/j.funbio.2017.10.007. Epub 2017 Oct 26. Fungal Biol. 2018. PMID: 29801792
-
Mating type-dependent partner sensing as mediated by VEL1 in Trichoderma reesei.Mol Microbiol. 2015 Jun;96(6):1103-18. doi: 10.1111/mmi.12993. Epub 2015 Apr 16. Mol Microbiol. 2015. PMID: 25757597 Free PMC article.
-
ENVOY is a major determinant in regulation of sexual development in Hypocrea jecorina (Trichoderma reesei).Eukaryot Cell. 2012 Jul;11(7):885-95. doi: 10.1128/EC.05321-11. Epub 2012 May 11. Eukaryot Cell. 2012. PMID: 22581525 Free PMC article.
-
Trichoderma: sensing the environment for survival and dispersal.Microbiology (Reading). 2012 Jan;158(Pt 1):3-16. doi: 10.1099/mic.0.052688-0. Epub 2011 Sep 29. Microbiology (Reading). 2012. PMID: 21964734 Review.
-
Regulation of plant cell wall degradation by light in Trichoderma.Fungal Biol Biotechnol. 2018 Apr 24;5:10. doi: 10.1186/s40694-018-0052-7. eCollection 2018. Fungal Biol Biotechnol. 2018. PMID: 29713489 Free PMC article. Review.
Cited by
-
Co-cultivation of Trichoderma asperellum GDFS1009 and Bacillus amyloliquefaciens 1841 Causes Differential Gene Expression and Improvement in the Wheat Growth and Biocontrol Activity.Front Microbiol. 2019 May 16;10:1068. doi: 10.3389/fmicb.2019.01068. eCollection 2019. Front Microbiol. 2019. PMID: 31156586 Free PMC article.
-
Gene regulation associated with sexual development and female fertility in different isolates of Trichoderma reesei.Fungal Biol Biotechnol. 2018 May 15;5:9. doi: 10.1186/s40694-018-0055-4. eCollection 2018. Fungal Biol Biotechnol. 2018. PMID: 29785273 Free PMC article.
-
New approaches in bioprocess-control: Consortium guidance by synthetic cell-cell communication based on fungal pheromones.Eng Life Sci. 2018 Apr 14;18(6):387-400. doi: 10.1002/elsc.201700181. eCollection 2018 Jun. Eng Life Sci. 2018. PMID: 32624919 Free PMC article. Review.
-
Molecular regulation of fungal secondary metabolism.World J Microbiol Biotechnol. 2023 May 20;39(8):204. doi: 10.1007/s11274-023-03649-6. World J Microbiol Biotechnol. 2023. PMID: 37209190 Review.
-
Adhesion as a Focus in Trichoderma-Root Interactions.J Fungi (Basel). 2022 Apr 6;8(4):372. doi: 10.3390/jof8040372. J Fungi (Basel). 2022. PMID: 35448603 Free PMC article. Review.
References
-
- Schmoll M, Seiboth B, Druzhinina I, Kubicek C. Genomics analysis of biocontrol species and industrial enzyme producers from the genus Trichoderma In: Nowrousian M, editor. Fungal Genomics. The Mycota. XIII Heidelberg, Germany: Springer; 2014. p. 233–65.
-
- Schmoll M, Wang TF. Sexual development in Trichoderma In: Wendland J, editor. The Mycota (Vol I): Growth, Differentiation and Sexuality. The Mycota. Switzerland: Springer International Publishing; 2016. p. 457–74.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

