Complete study demonstrating the absence of rhabdovirus in a distinct Sf9 cell line
- PMID: 28423032
- PMCID: PMC5397025
- DOI: 10.1371/journal.pone.0175633
Complete study demonstrating the absence of rhabdovirus in a distinct Sf9 cell line
Abstract
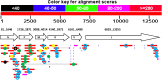
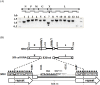
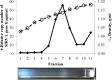
A putative novel rhabdovirus (SfRV) was previously identified in a Spodoptera frugiperda cell line (Sf9 cells [ATCC CRL-1711 lot 58078522]) by next generation sequencing and extensive bioinformatic analysis. We performed an extensive analysis of our Sf9 cell bank (ATCC CRL-1711 lot 5814 [Sf9L5814]) to determine whether this virus was already present in cells obtained from ATCC in 1987. Inverse PCR of DNA isolated from Sf9 L5814 cellular DNA revealed integration of SfRV sequences in the cellular genome. RT-PCR of total RNA showed a deletion of 320 nucleotides in the SfRV RNA that includes the transcriptional motifs for genes X and L. Concentrated cell culture supernatant was analyzed by sucrose density gradient centrifugation and revealed a single band at a density of 1.14 g/ml. This fraction was further analysed by electron microscopy and showed amorphous and particulate debris that did not resemble a rhabdovirus in morphology or size. SDS-PAGE analysis confirmed that the protein composition did not contain the typical five rhabdovirus structural proteins and LC-MS/MS analysis revealed primarily of exosomal marker proteins, the SfRV N protein, and truncated forms of SfRV N, P, and G proteins. The SfRV L gene fragment RNA sequence was recovered from the supernatant after ultracentrifugation of the 1.14 g/ml fraction treated with diethyl ether suggesting that the SfRV L gene fragment sequence is not associated with a diethyl ether resistant nucleocapsid. Interestingly, the 1.14 g/ml fraction was able to transfer baculovirus DNA into Sf9L5814 cells, consistent with the presence of functional exosomes. Our results demonstrate the absence of viral particles in ATCC CRL-1711 lot 5814 Sf9 cells in contrast to a previous study that suggested the presence of infectious rhabdoviral particles in Sf9 cells from a different lot. This study highlights how cell lines with different lineages may present different virosomes and therefore no general conclusions can be drawn across Sf9 cells from different laboratories.
Conflict of interest statement
Figures



Similar articles
-
Long-Read High-Throughput Sequencing (HTS) Revealed That the Sf-Rhabdovirus X+ Genome Contains a 3.7 kb Internal Duplication.Viruses. 2023 Sep 26;15(10):1998. doi: 10.3390/v15101998. Viruses. 2023. PMID: 37896775 Free PMC article.
-
Identification of a novel rhabdovirus in Spodoptera frugiperda cell lines.J Virol. 2014 Jun;88(12):6576-85. doi: 10.1128/JVI.00780-14. Epub 2014 Mar 26. J Virol. 2014. PMID: 24672045 Free PMC article.
-
The Spodoptera frugiperda Sf9 cell line is a heterogeneous population of rhabdovirus-infected and virus-negative cells: Isolation and characterization of cell clones containing rhabdovirus X-gene variants and virus-negative cell clones.Virology. 2019 Oct;536:125-133. doi: 10.1016/j.virol.2019.08.001. Epub 2019 Aug 2. Virology. 2019. PMID: 31494355
-
Persistent Spodoptera frugiperda rhabdovirus infection in Sf9 cells is not restricted by Wolbachia wMelPop-CLA and wAlbB strains and is targeted by the RNAi machinery.Virology. 2021 Nov;563:82-87. doi: 10.1016/j.virol.2021.08.013. Epub 2021 Sep 2. Virology. 2021. PMID: 34492433
-
Knockout of Dicer-2 in the Sf9 cell line enhances the replication of Spodoptera frugiperda rhabdovirus and conditionally increases baculovirus replication.J Gen Virol. 2022 Aug;103(8). doi: 10.1099/jgv.0.001779. J Gen Virol. 2022. PMID: 36018884
Cited by
-
A database of crop pest cell lines.In Vitro Cell Dev Biol Anim. 2022 Sep;58(8):719-757. doi: 10.1007/s11626-022-00710-w. Epub 2022 Aug 22. In Vitro Cell Dev Biol Anim. 2022. PMID: 35994130
-
Characterisation of extracellular vesicles in baculovirus infection of Spodoptera frugiperda cells.J Extracell Biol. 2024 Jun 28;3(7):e163. doi: 10.1002/jex2.163. eCollection 2024 Jul. J Extracell Biol. 2024. PMID: 38947876 Free PMC article.
-
Long-Read High-Throughput Sequencing (HTS) Revealed That the Sf-Rhabdovirus X+ Genome Contains a 3.7 kb Internal Duplication.Viruses. 2023 Sep 26;15(10):1998. doi: 10.3390/v15101998. Viruses. 2023. PMID: 37896775 Free PMC article.
-
Expressional Localization and Functionally Identifying an RNA Editing Enzyme BmADARa of the Silkworm Bombyx mori.Insects. 2020 Aug 12;11(8):523. doi: 10.3390/insects11080523. Insects. 2020. PMID: 32806497 Free PMC article.
-
Infectivity of Sf-rhabdovirus variants in insect and mammalian cell lines.Virology. 2017 Dec;512:234-245. doi: 10.1016/j.virol.2017.09.025. Epub 2017 Oct 9. Virology. 2017. PMID: 29024851 Free PMC article.
References
-
- Ma H, Galvin T a, Glasner DR, Shaheduzzaman S, Khan AS. Identification of a novel rhabdovirus in Spodoptera frugiperda cell lines. J Virol. 2014;88: 6576–85. doi: 10.1128/JVI.00780-14 - DOI - PMC - PubMed
-
- Cox MMJ, Izikson R, Post P, Dunkle L. Safety, efficacy, and immunogenicity of Flublok in the prevention of seasonal influenza in adults. Ther Adv vaccines. 2015;3: 97–108. doi: 10.1177/2051013615595595 - DOI - PMC - PubMed
-
- McPherson CE. Development of a novel recombinant influenza vaccine in insect cells. Biologicals. The International Association for Biologicals; 2008;36: 350–353. doi: 10.1016/j.biologicals.2008.08.001 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

