Biochemical and molecular characterization of the isocitrate dehydrogenase with dual coenzyme specificity from the obligate methylotroph Methylobacillus Flagellatus
- PMID: 28423051
- PMCID: PMC5397045
- DOI: 10.1371/journal.pone.0176056
Biochemical and molecular characterization of the isocitrate dehydrogenase with dual coenzyme specificity from the obligate methylotroph Methylobacillus Flagellatus
Abstract
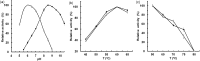
The isocitrate dehydrogenase (MfIDH) with unique double coenzyme specificity from Methylobacillus flagellatus was purified and characterized, and its gene was cloned and overexpressed in E. coli as a fused protein. This enzyme is homodimeric,-with a subunit molecular mass of 45 kDa and a specific activity of 182 U mg -1 with NAD+ and 63 U mg -1 with NADP+. The MfIDH activity was dependent on divalent cations and Mn2+ enhanced the activity the most effectively. MfIDH exhibited a cofactor-dependent pH-activity profile. The optimum pH values were 8.5 (NAD+) and 6.0 (NADP+).The Km values for NAD+ and NADP+ were 113 μM and 184 μM respectively, while the Km values for DL-isocitrate were 9.0 μM (NAD+), 8.0 μM (NADP+). The MfIDH specificity (kcat/Km) was only 5-times higher for NAD+ than for NADP+. The purified MfIDH displayed maximal activity at 60°C. Heat-inactivation studies showed that the MfIDH was remarkably thermostable, retaining full activity at 50°C and losting ca. 50% of its activity after one hour of incubation at 75°C. The enzyme was insensitive to the presence of intermediate metabolites, with the exception of 2 mM ATP, which caused 50% inhibition of NADP+-linked activity. The indispensability of the N6 amino group of NAD(P)+ in its binding to MfIDH was demonstrated. MfIDH showed high sequence similarity with bacterial NAD(P)+-dependent type I isocitrate dehydrogenases (IDHs) rather than with eukaryotic NAD+-dependent IDHs. The unique double coenzyme specificity of MfIDH potentially resulted from the Lys340, Ile341 and Ala347 residues in the coenzyme-binding site of the enzyme. The discovery of a type I IDH with double coenzyme specificity elucidates the evolution of this subfamily IDHs and may provide fundamental information for engineering enzymes with desired properties.
Conflict of interest statement
Figures


Similar articles
-
A Novel Type II NAD+-Specific Isocitrate Dehydrogenase from the Marine Bacterium Congregibacter litoralis KT71.PLoS One. 2015 May 5;10(5):e0125229. doi: 10.1371/journal.pone.0125229. eCollection 2015. PLoS One. 2015. PMID: 25942017 Free PMC article.
-
Biochemical Characterization and Complete Conversion of Coenzyme Specificity of Isocitrate Dehydrogenase from Bifidobacterium longum.Int J Mol Sci. 2016 Feb 26;17(3):296. doi: 10.3390/ijms17030296. Int J Mol Sci. 2016. PMID: 26927087 Free PMC article.
-
NADP(+)-specific isocitrate dehydrogenase from oleaginous yeast Yarrowia lipolytica CLIB122: biochemical characterization and coenzyme sites evaluation.Appl Biochem Biotechnol. 2013 Sep;171(2):403-16. doi: 10.1007/s12010-013-0373-1. Epub 2013 Jul 12. Appl Biochem Biotechnol. 2013. PMID: 23846800
-
Glutamate dehydrogenases: the why and how of coenzyme specificity.Neurochem Res. 2014;39(3):426-32. doi: 10.1007/s11064-013-1089-x. Epub 2013 Jun 13. Neurochem Res. 2014. PMID: 23761034 Review.
-
Molecular properties, functions, and potential applications of NAD kinases.Acta Biochim Biophys Sin (Shanghai). 2009 May;41(5):352-61. doi: 10.1093/abbs/gmp029. Acta Biochim Biophys Sin (Shanghai). 2009. PMID: 19430699 Review.
Cited by
-
Dysfunctional cardiac energy transduction, mitochondrial oxidative stress, oncogenic and apoptotic signaling in DiNP-induced asthma in murine model.Naunyn Schmiedebergs Arch Pharmacol. 2025 Mar;398(3):2833-2843. doi: 10.1007/s00210-024-03454-4. Epub 2024 Sep 17. Naunyn Schmiedebergs Arch Pharmacol. 2025. PMID: 39287675 Free PMC article.
-
Impaired energy metabolism and altered brain histoarchitecture characterized by inhibition of glycolysis and mitochondrial electron transport-linked enzymes in rats exposed to diisononyl phthalate.Heliyon. 2024 Aug 10;10(16):e36056. doi: 10.1016/j.heliyon.2024.e36056. eCollection 2024 Aug 30. Heliyon. 2024. Retraction in: Heliyon. 2025 Mar 12;11(6):e43159. doi: 10.1016/j.heliyon.2025.e43159. PMID: 39224312 Free PMC article. Retracted.
-
Assessment of neuro-pulmonary crosstalk in asthmatic mice: effects of DiNP exposure on cellular respiration, mitochondrial oxidative status and apoptotic signaling.Sci Rep. 2024 Jun 26;14(1):14712. doi: 10.1038/s41598-024-65356-y. Sci Rep. 2024. PMID: 38926453 Free PMC article.
-
Enzymatic Characterization of the Isocitrate Dehydrogenase with Dual Coenzyme Specificity from the Marine Bacterium Umbonibacter marinipuiceus.Int J Mol Sci. 2023 Jul 13;24(14):11428. doi: 10.3390/ijms241411428. Int J Mol Sci. 2023. PMID: 37511187 Free PMC article.
References
-
- Zhu G, Golding GB, Dean AM. The selective cause of an ancient adaptation. Science. 2005;307: 1279–1282. doi: 10.1126/science.1106974 - DOI - PubMed
-
- Shimizu T, Yin L, Yoshida A, Yokooji Y, Hachisuka S, Sato T, et al. Structure and function of an ancestral-type β-decarboxylating dehydrogenase from Thermococcus kodakarensis. Biochem J. 2016; 105–122. doi: 10.1042/BCJ20160699 - DOI - PubMed
-
- Wu M-C, Tian C-Q, Cheng H-M, Xu L, Wang P, Zhu G-P. A novel type II NAD+-specific isocitrate dehydrogenase from the marine bacterium Congregibacter litoralis KT71. PLoS One. 2015;10: e0125229 doi: 10.1371/journal.pone.0125229 - DOI - PMC - PubMed
-
- Banerjee S, Nandyala A, Podili R, Katoch VM, Hasnain SE. Comparison of Mycobacterium tuberculosis isocitrate dehydrogenases (ICD-1 and ICD-2) reveals differences in coenzyme affinity, oligomeric state, pH tolerance and phylogenetic affiliation. BMC Biochem. 2005;6: 20 doi: 10.1186/1471-2091-6-20 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

