Hepatocellular Carcinomas Originate Predominantly from Hepatocytes and Benign Lesions from Hepatic Progenitor Cells
- PMID: 28423321
- PMCID: PMC5409928
- DOI: 10.1016/j.celrep.2017.03.059
Hepatocellular Carcinomas Originate Predominantly from Hepatocytes and Benign Lesions from Hepatic Progenitor Cells
Abstract
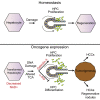
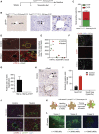
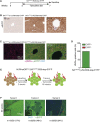
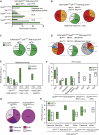
Hepatocellular carcinoma (HCC) is an aggressive primary liver cancer. However, its origin remains a debated question. Using human data and various hepatocarcinogenesis mouse models, we show that, in early stages, transformed hepatocytes, independent of their proliferation status, activate hepatic progenitor cell (HPC) expansion. Genetic lineage tracing of HPCs and hepatocytes reveals that, in all models, HCC originates from hepatocytes. However, whereas in various models tumors do not emanate from HPCs, tracking of progenitors in a model mimicking human hepatocarcinogenesis indicates that HPCs can generate benign lesions (regenerative nodules and adenomas) and aggressive HCCs. Mechanistically, galectin-3 and α-ketoglutarate paracrine signals emanating from oncogene-expressing hepatocytes instruct HPCs toward HCCs. α-Ketoglutarate preserves an HPC undifferentiated state, and galectin-3 maintains HPC stemness, expansion, and aggressiveness. Pharmacological or genetic blockage of galectin-3 reduces HCC, and its expression in human HCC correlates with poor survival. Our findings may have clinical implications for liver regeneration and HCC therapy.
Keywords: DNA damage; HCC; NAD(+); adenomas; galectin-3; hepatic progenitor cells; hepatocytes; lineage tracking; regenerative nodules; α-ketoglutarate.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Boyault S., Rickman D.S., de Reyniès A., Balabaud C., Rebouissou S., Jeannot E., Hérault A., Saric J., Belghiti J., Franco D. Transcriptome classification of HCC is related to gene alterations and to new therapeutic targets. Hepatology. 2007;45:42–52. - PubMed
-
- Burén S., Gomes A.L., Teijeiro A., Fawal M.A., Yilmaz M., Tummala K.S., Perez M., Rodriguez-Justo M., Campos-Olivas R., Megías D., Djouder N. Regulation of OGT by URI in Response to Glucose Confers c-MYC-Dependent Survival Mechanisms. Cancer Cell. 2016;30:290–307. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

