The PIAS3-Smurf2 sumoylation pathway suppresses breast cancer organoid invasiveness
- PMID: 28423498
- PMCID: PMC5400561
- DOI: 10.18632/oncotarget.15471
The PIAS3-Smurf2 sumoylation pathway suppresses breast cancer organoid invasiveness
Abstract
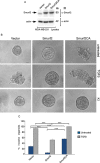
Tumor metastasis profoundly reduces the survival of breast cancer patients, but the mechanisms underlying breast cancer invasiveness and metastasis are incompletely understood. Here, we report that the E3 ubiquitin ligase Smurf2 acts in a sumoylation-dependent manner to suppress the invasive behavior of MDA-MB-231 human breast cancer cell-derived organoids. We also find that the SUMO E3 ligase PIAS3 inhibits the invasive growth of breast cancer cell-derived organoids. In mechanistic studies, PIAS3 maintains breast cancer organoids in a non-invasive state via sumoylation of Smurf2. Importantly, the E3 ubiquitin ligase activity is required for sumoylated Smurf2 to suppress the invasive growth of breast cancer-cell derived organoids. Collectively, our findings define a novel role for the PIAS3-Smurf2 sumoylation pathway in the suppression of breast cancer cell invasiveness. These findings lay the foundation for the development of novel biomarkers and targeted therapeutic approaches in breast cancer.
Keywords: breast cancer; invasion; sumoylation.
Conflict of interest statement
None.
Figures
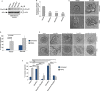
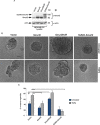

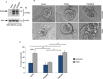
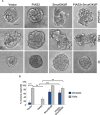


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

