The novel long non-coding RNA TALNEC2, regulates tumor cell growth and the stemness and radiation response of glioma stem cells
- PMID: 28423669
- PMCID: PMC5458248
- DOI: 10.18632/oncotarget.15991
The novel long non-coding RNA TALNEC2, regulates tumor cell growth and the stemness and radiation response of glioma stem cells
Erratum in
-
Correction: The novel long non-coding RNA TALNEC2, regulates tumor cell growth and the stemness and radiation response of glioma stem cells.Oncotarget. 2021 Dec 21;12(26):2546-2547. doi: 10.18632/oncotarget.27383. eCollection 2021 Dec 21. Oncotarget. 2021. PMID: 34966487 Free PMC article.
Abstract
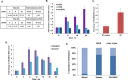
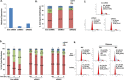
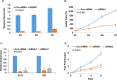
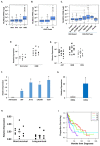
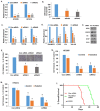
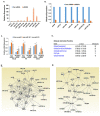
Despite advances in novel therapeutic approaches for the treatment of glioblastoma (GBM), the median survival of 12-14 months has not changed significantly. Therefore, there is an imperative need to identify molecular mechanisms that play a role in patient survival. Here, we analyzed the expression and functions of a novel lncRNA, TALNEC2 that was identified using RNA seq of E2F1-regulated lncRNAs. TALNEC2 was localized to the cytosol and its expression was E2F1-regulated and cell-cycle dependent. TALNEC2 was highly expressed in GBM with poor prognosis, in GBM specimens derived from short-term survivors and in glioma cells and glioma stem cells (GSCs). Silencing of TALNEC2 inhibited cell proliferation and arrested the cells in the G1\S phase of the cell cycle in various cancer cell lines. In addition, silencing of TALNEC2 decreased the self-renewal and mesenchymal transformation of GSCs, increased sensitivity of these cells to radiation and prolonged survival of mice bearing GSC-derived xenografts. Using miRNA array analysis, we identified specific miRNAs that were altered in the silenced cells that were associated with cell-cycle progression, proliferation and mesenchymal transformation. Two of the downregulated miRNAs, miR-21 and miR-191, mediated some of TALNEC2 effects on the stemness and mesenchymal transformation of GSCs. In conclusion, we identified a novel E2F1-regulated lncRNA that is highly expressed in GBM and in tumors from patients of short-term survival. The expression of TALNEC2 is associated with the increased tumorigenic potential of GSCs and their resistance to radiation. We conclude that TALNEC2 is an attractive therapeutic target for the treatment of GBM.
Keywords: TALNEC2; glioblastoma; glioma stem cells; long non-cording RNAs; mesenchymal transformation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
NONHSAT141192.2 Facilitates the Stemness and Radioresistance of Glioma Stem Cells via the Regulation of PIK3R3 and SOX2.CNS Neurosci Ther. 2025 Feb;31(2):e70279. doi: 10.1111/cns.70279. CNS Neurosci Ther. 2025. PMID: 39968701 Free PMC article.
-
Mir-370-3p Impairs Glioblastoma Stem-Like Cell Malignancy Regulating a Complex Interplay between HMGA2/HIF1A and the Oncogenic Long Non-Coding RNA (lncRNA) NEAT1.Int J Mol Sci. 2020 May 20;21(10):3610. doi: 10.3390/ijms21103610. Int J Mol Sci. 2020. PMID: 32443824 Free PMC article.
-
Long non-coding RNA lung cancer-associated transcript-1 promotes glioblastoma progression by enhancing Hypoxia-inducible factor 1 alpha activity.Neuro Oncol. 2024 Aug 5;26(8):1388-1401. doi: 10.1093/neuonc/noae036. Neuro Oncol. 2024. PMID: 38456228 Free PMC article.
-
Integrative analysis of cell adhesion molecules in glioblastoma identified prostaglandin F2 receptor inhibitor (PTGFRN) as an essential gene.BMC Cancer. 2022 Jun 11;22(1):642. doi: 10.1186/s12885-022-09682-2. BMC Cancer. 2022. PMID: 35690717 Free PMC article. Review.
-
Functional Roles of Long Non-Coding RNAs (LncRNAs) in Glioma Stem Cells.Med Sci Monit. 2019 Oct 8;25:7567-7573. doi: 10.12659/MSM.916040. Med Sci Monit. 2019. PMID: 31593561 Free PMC article. Review.
Cited by
-
Crosstalk between SOX Genes and Long Non-Coding RNAs in Glioblastoma.Int J Mol Sci. 2023 Mar 28;24(7):6392. doi: 10.3390/ijms24076392. Int J Mol Sci. 2023. PMID: 37047365 Free PMC article. Review.
-
Exploring the Roles of lncRNAs in GBM Pathophysiology and Their Therapeutic Potential.Cells. 2020 Oct 28;9(11):2369. doi: 10.3390/cells9112369. Cells. 2020. PMID: 33126510 Free PMC article. Review.
-
Genomic and Epigenomic Features of Glioblastoma Multiforme and its Biomarkers.J Oncol. 2022 Sep 21;2022:4022960. doi: 10.1155/2022/4022960. eCollection 2022. J Oncol. 2022. PMID: 36185622 Free PMC article. Review.
-
PR-LncRNA signature regulates glioma cell activity through expression of SOX factors.Sci Rep. 2018 Aug 24;8(1):12746. doi: 10.1038/s41598-018-30836-5. Sci Rep. 2018. PMID: 30143669 Free PMC article.
-
The long non-coding RNA SNHG14 inhibits cell proliferation and invasion and promotes apoptosis by sponging miR-92a-3p in glioma.Oncotarget. 2018 Jan 4;9(15):12112-12124. doi: 10.18632/oncotarget.23960. eCollection 2018 Feb 23. Oncotarget. 2018. PMID: 29552296 Free PMC article.
References
-
- Brunner AL, Beck AH, Edris B, Sweeney RT, Zhu SX, Li R, Montgomery K, Varma S, Gilks T, Guo X, Foley JW, Witten DM, Giacomini CP, et al. Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome biology. 2012;13:R75. - PMC - PubMed
-
- Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

