RMP promotes epithelial-mesenchymal transition through NF-κB/CSN2/Snail pathway in hepatocellular carcinoma
- PMID: 28423737
- PMCID: PMC5522250
- DOI: 10.18632/oncotarget.16177
RMP promotes epithelial-mesenchymal transition through NF-κB/CSN2/Snail pathway in hepatocellular carcinoma
Abstract
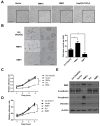
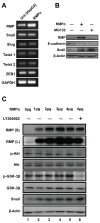
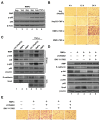
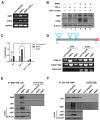
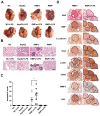
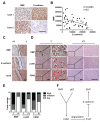
Epithelial-mesenchymal transition (EMT) is a significant risk factor for metastasis in hepatocellular carcinoma (HCC) patients and with poor prognosis. In this study, we demonstrate the key role of RPB5-mediating protein (RMP) in EMT of HCC cells and the mechanism by which RMP promote EMT. RMP increases migration, invasion, and the progress of EMT of HCC cells, which facilitates the accumulation of Snail, a transcriptional repressor involved in EMT initiation. NF-κB is activated by RMP, which directly promotes the expression of COP9 signalosome 2 (CSN2) to repress the degradation of Snail. Pulmonary metastases mouse model demonstrates that RMP induces metastasis in vivo. Immunohistochemical analysis of human HCC tissues confirms the correlation of RMP with the expression of E-cadherin, p65, CSN2 and Snail in vivo. Collectively, these findings indicate that RMP promotes EMT and HCC metastasis through NF-κB/CSN2/Snail pathway. These results suggest that RMP and p65 may serve as potential candidates of the targets in the treatment of metastatic HCC.
Keywords: EMT; HCC; NF-κB; RMP; metastasis.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article.
Figures



Similar articles
-
Post-transcription mediated Snail stabilization is involved in radiation exposure induced invasion and migration of hepatocarcinoma cells.Biomed Pharmacother. 2018 Jul;103:767-772. doi: 10.1016/j.biopha.2018.04.095. Epub 2018 Apr 24. Biomed Pharmacother. 2018. PMID: 29684855
-
Overexpression of CSN6 promotes the epithelial-mesenchymal transition and predicts poor prognosis in hepatocellular carcinoma.Clin Res Hepatol Gastroenterol. 2020 Jun;44(3):340-348. doi: 10.1016/j.clinre.2019.07.012. Epub 2019 Aug 14. Clin Res Hepatol Gastroenterol. 2020. PMID: 31422034
-
FoxM1 overexpression promotes epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma.World J Gastroenterol. 2015 Jan 7;21(1):196-213. doi: 10.3748/wjg.v21.i1.196. World J Gastroenterol. 2015. PMID: 25574092 Free PMC article.
-
The novel role of Yin Yang 1 in the regulation of epithelial to mesenchymal transition in cancer via the dysregulated NF-κB/Snail/YY1/RKIP/PTEN Circuitry.Crit Rev Oncog. 2011;16(3-4):211-26. doi: 10.1615/critrevoncog.v16.i3-4.50. Crit Rev Oncog. 2011. PMID: 22248055 Review.
-
Re-exploration of immunotherapy targeting EMT of hepatocellular carcinoma: Starting from the NF-κB pathway.Biomed Pharmacother. 2024 May;174:116566. doi: 10.1016/j.biopha.2024.116566. Epub 2024 Apr 16. Biomed Pharmacother. 2024. PMID: 38631143 Review.
Cited by
-
Role of tight junction-associated MARVEL protein marvelD3 in migration and epithelial-mesenchymal transition of hepatocellular carcinoma.Cell Adh Migr. 2021 Dec;15(1):249-260. doi: 10.1080/19336918.2021.1958441. Cell Adh Migr. 2021. PMID: 34338154 Free PMC article.
-
RMP promotes the proliferation and radioresistance of esophageal carcinoma.J Cancer. 2019 Jun 9;10(16):3698-3705. doi: 10.7150/jca.32680. eCollection 2019. J Cancer. 2019. PMID: 31333787 Free PMC article.
-
RMP/URI inhibits both intrinsic and extrinsic apoptosis through different signaling pathways.Int J Biol Sci. 2019 Oct 15;15(12):2692-2706. doi: 10.7150/ijbs.36829. eCollection 2019. Int J Biol Sci. 2019. PMID: 31754340 Free PMC article.
-
Hypoxia promotes metastasis by relieving miR-598-3p-restricted glycolysis in gastric cancer.J Transl Med. 2024 Mar 15;22(1):283. doi: 10.1186/s12967-024-04957-7. J Transl Med. 2024. PMID: 38491378 Free PMC article.
-
The Role of microRNAs in Embryonic and Induced Pluripotency.J Stem Cells Regen Med. 2018 May 30;14(1):3-9. doi: 10.46582/jsrm.1401002. eCollection 2018. J Stem Cells Regen Med. 2018. PMID: 30018468 Free PMC article. Review.
References
-
- Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. - PubMed
-
- Budhu A, Forgues M, Ye QH, Jia HL, He P, Zanetti KA, Kammula US, Chen YD, Qin LX, Tang ZY, Wang XW. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111. - PubMed
-
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nature reviews Cancer. 2002;2:442–454. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

