Plasma metabolomics for the diagnosis and prognosis of H1N1 influenza pneumonia
- PMID: 28424077
- PMCID: PMC5397800
- DOI: 10.1186/s13054-017-1672-7
Plasma metabolomics for the diagnosis and prognosis of H1N1 influenza pneumonia
Abstract
Background: Metabolomics is a tool that has been used for the diagnosis and prognosis of specific diseases. The purpose of this study was to examine if metabolomics could be used as a potential diagnostic and prognostic tool for H1N1 pneumonia. Our hypothesis was that metabolomics can potentially be used early for the diagnosis and prognosis of H1N1 influenza pneumonia.
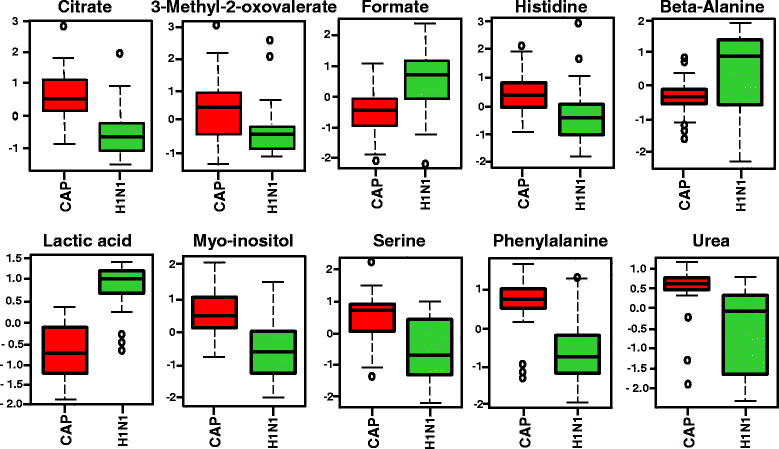
Methods: 1H nuclear magnetic resonance spectroscopy and gas chromatography-mass spectrometry were used to profile the metabolome in 42 patients with H1N1 pneumonia, 31 ventilated control subjects in the intensive care unit (ICU), and 30 culture-positive plasma samples from patients with bacterial community-acquired pneumonia drawn within the first 24 h of hospital admission for diagnosis and prognosis of disease.
Results: We found that plasma-based metabolomics from samples taken within 24 h of hospital admission can be used to discriminate H1N1 pneumonia from bacterial pneumonia and nonsurvivors from survivors of H1N1 pneumonia. Moreover, metabolomics is a highly sensitive and specific tool for the 90-day prognosis of mortality in H1N1 pneumonia.
Conclusions: This study demonstrates that H1N1 pneumonia can create a quite different plasma metabolic profile from bacterial culture-positive pneumonia and ventilated control subjects in the ICU on the basis of plasma samples taken within 24 h of hospital/ICU admission, early in the course of disease.
Keywords: Biomarkers; GC-MS; H1N1 pneumonia; Metabolomics; NMR.
Figures





References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

