The within-host population dynamics of Mycobacterium tuberculosis vary with treatment efficacy
- PMID: 28424085
- PMCID: PMC5395877
- DOI: 10.1186/s13059-017-1196-0
The within-host population dynamics of Mycobacterium tuberculosis vary with treatment efficacy
Abstract
Background: Combination therapy is one of the most effective tools for limiting the emergence of drug resistance in pathogens. Despite the widespread adoption of combination therapy across diseases, drug resistance rates continue to rise, leading to failing treatment regimens. The mechanisms underlying treatment failure are well studied, but the processes governing successful combination therapy are poorly understood. We address this question by studying the population dynamics of Mycobacterium tuberculosis within tuberculosis patients undergoing treatment with different combinations of antibiotics.
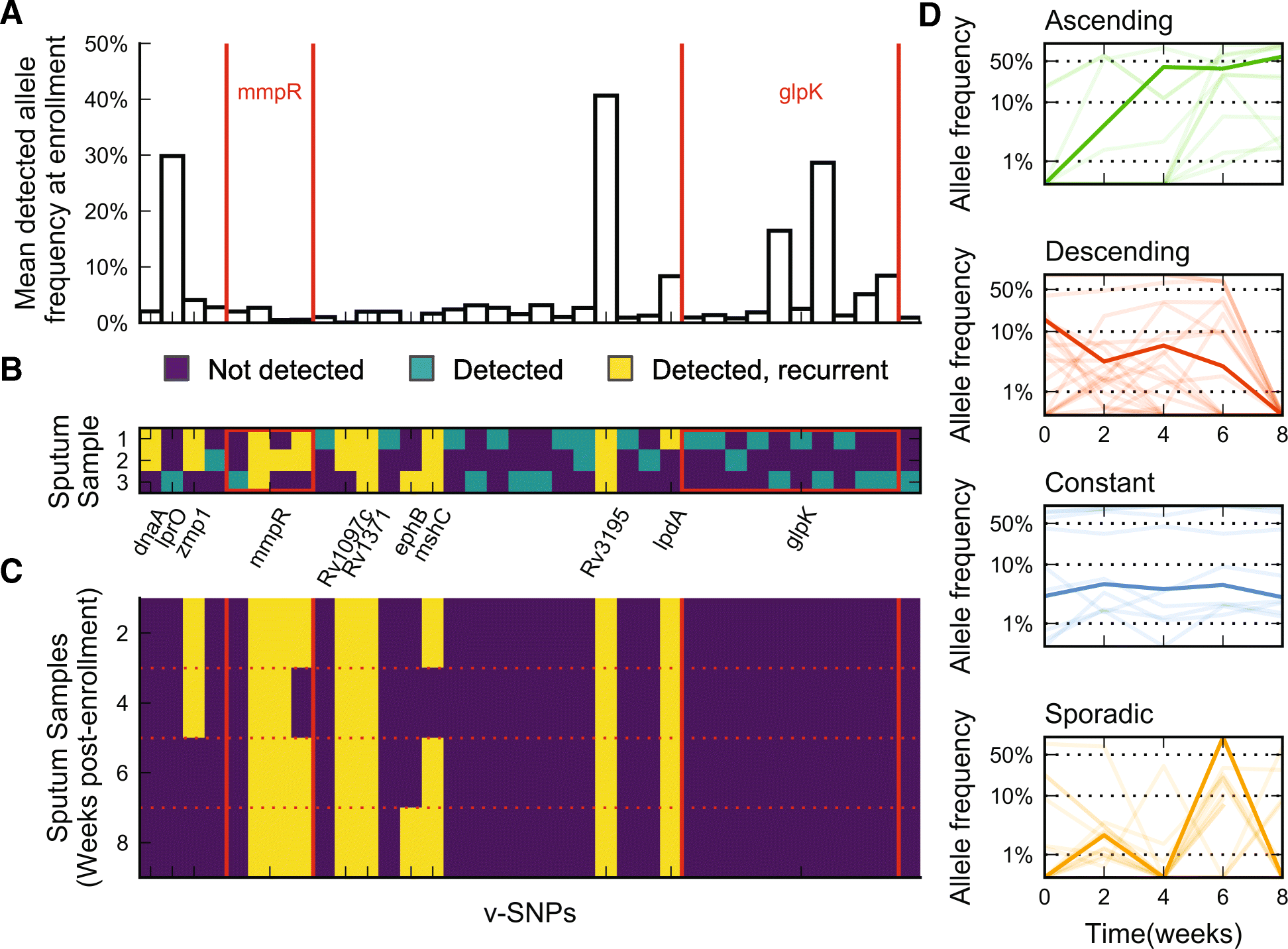
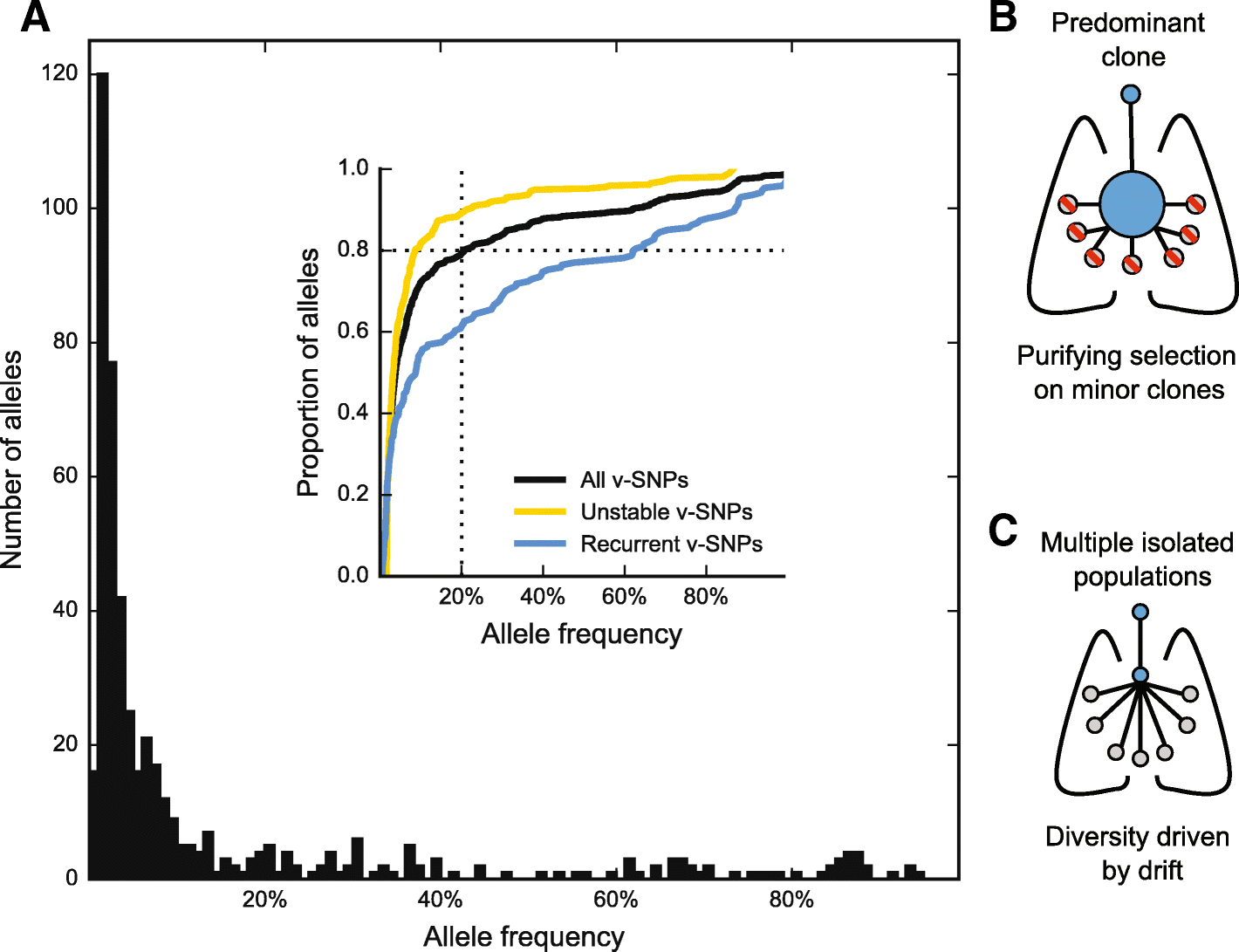
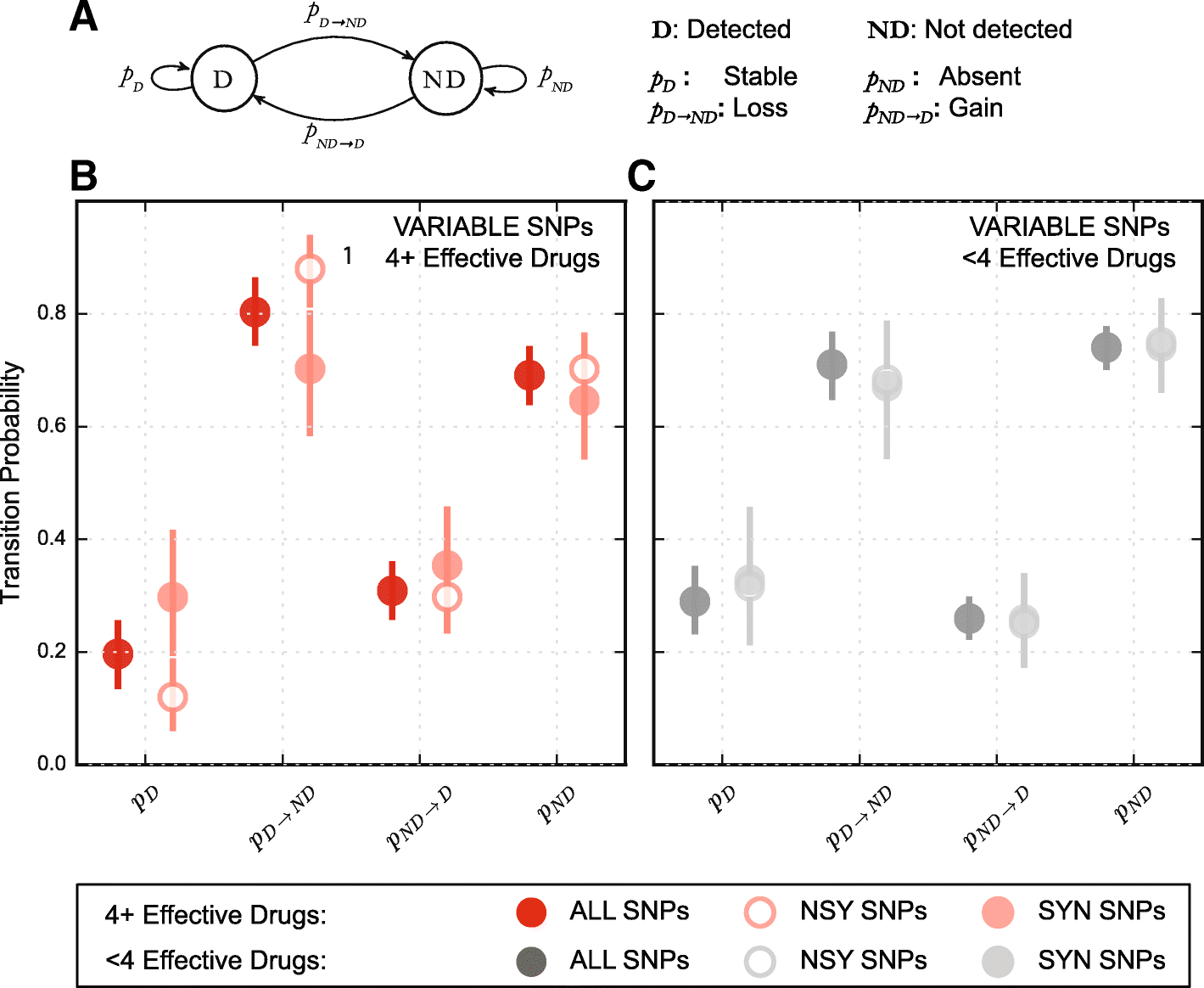
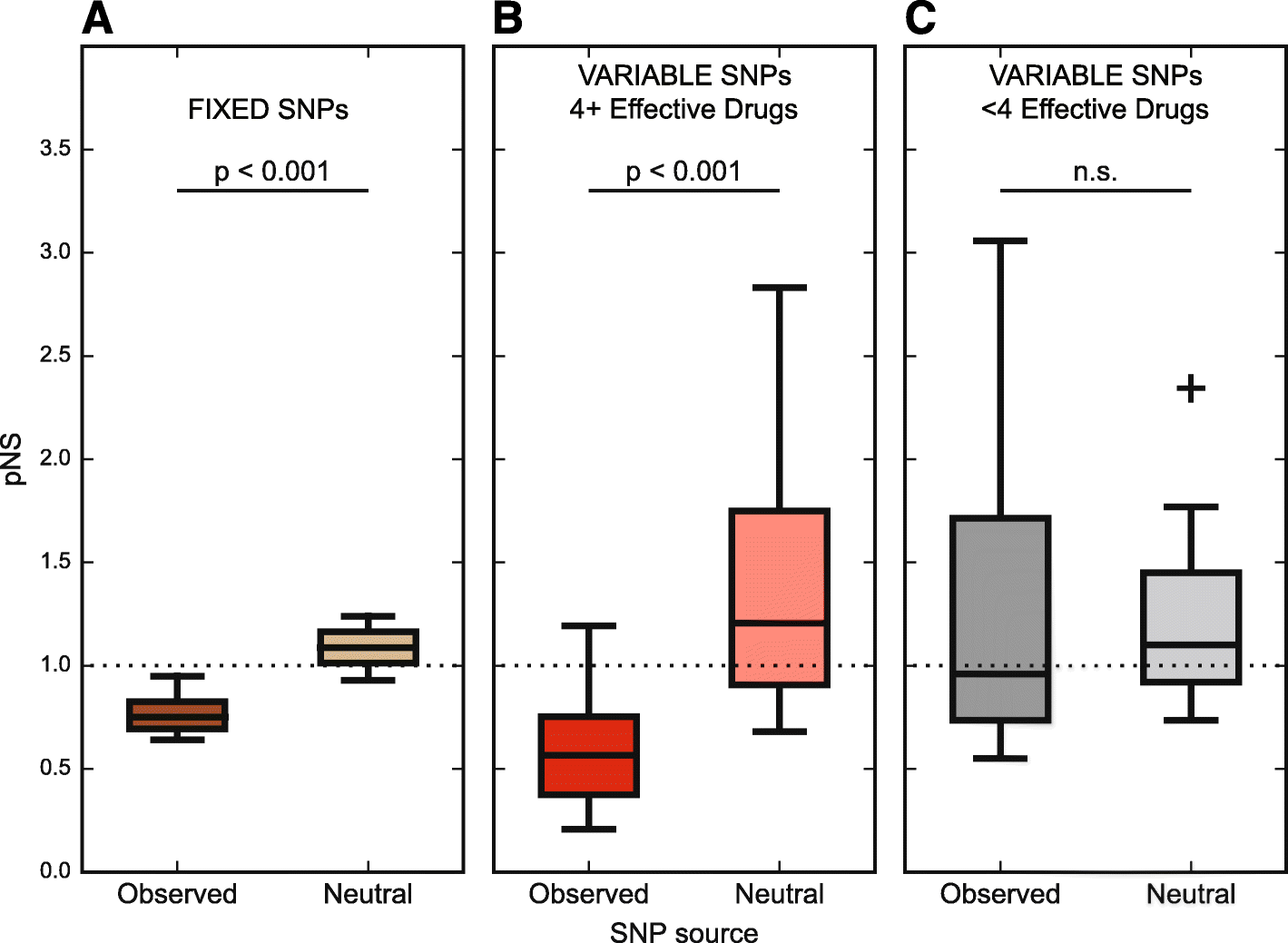
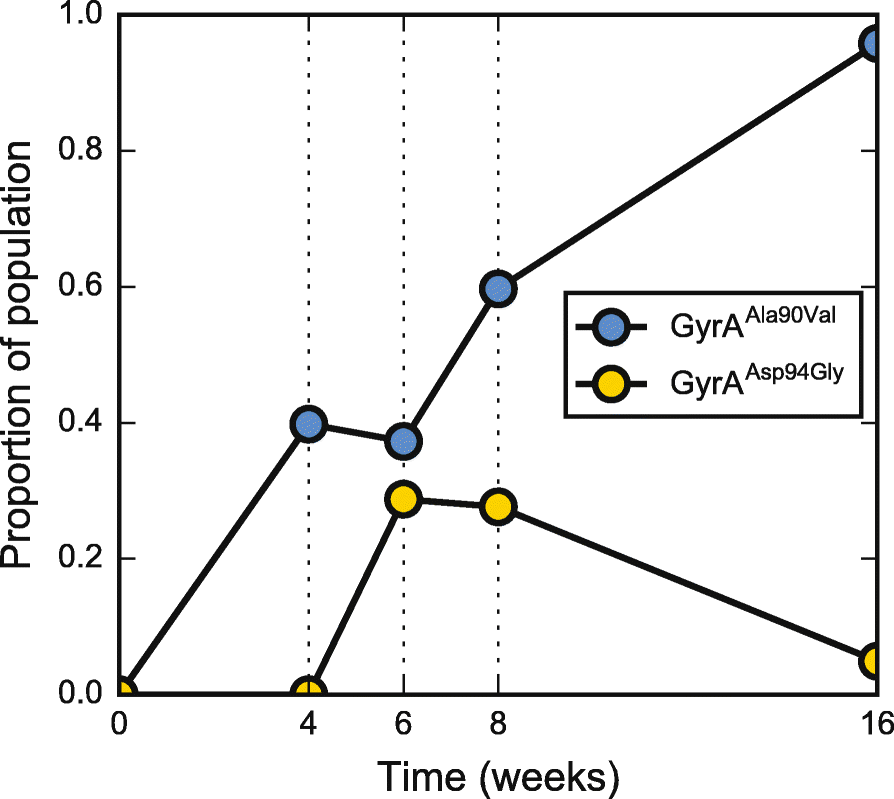
Results: By combining very deep whole genome sequencing (~1000-fold genome-wide coverage) with sequential sputum sampling, we were able to detect transient genetic diversity driven by the apparently continuous turnover of minor alleles, which could serve as the source of drug-resistant bacteria. However, we report that treatment efficacy has a clear impact on the population dynamics: sufficient drug pressure bears a clear signature of purifying selection leading to apparent genetic stability. In contrast, M. tuberculosis populations subject to less drug pressure show markedly different dynamics, including cases of acquisition of additional drug resistance.
Conclusions: Our findings show that for a pathogen like M. tuberculosis, which is well adapted to the human host, purifying selection constrains the evolutionary trajectory to resistance in effectively treated individuals. Nonetheless, we also report a continuous turnover of minor variants, which could give rise to the emergence of drug resistance in cases of drug pressure weakening. Monitoring bacterial population dynamics could therefore provide an informative metric for assessing the efficacy of novel drug combinations.
Trial registration: ClinicalTrials.gov NCT01071603.
Keywords: Combination therapy; Drug resistance; Tuberculosis; Whole genome sequencing; Within-host evolution.
Figures

Comment in
-
The path of least antibiotic resistance.Sci Transl Med. 2017 May 10;9(389):eaan3778. doi: 10.1126/scitranslmed.aan3778. Sci Transl Med. 2017. PMID: 28490666
References
-
- Fox W, Ellard GA, Mitchison DA. Studies on the treatment of tuberculosis undertaken by the British Medical Research Council tuberculosis units, 1946-1986, with relevant subsequent publications. Int J Tuberc Lung Dis. 1999;3:S231–79. - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

