Subtype-Specific Differences in Gag-Protease-Driven Replication Capacity Are Consistent with Intersubtype Differences in HIV-1 Disease Progression
- PMID: 28424286
- PMCID: PMC5469260
- DOI: 10.1128/JVI.00253-17
Subtype-Specific Differences in Gag-Protease-Driven Replication Capacity Are Consistent with Intersubtype Differences in HIV-1 Disease Progression
Abstract
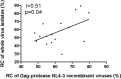
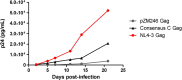
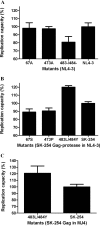
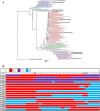
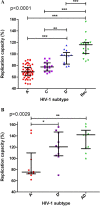
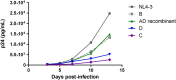
There are marked differences in the spread and prevalence of HIV-1 subtypes worldwide, and differences in clinical progression have been reported. However, the biological reasons underlying these differences are unknown. Gag-protease is essential for HIV-1 replication, and Gag-protease-driven replication capacity has previously been correlated with disease progression. We show that Gag-protease replication capacity correlates significantly with that of whole isolates (r = 0.51; P = 0.04), indicating that Gag-protease is a significant contributor to viral replication capacity. Furthermore, we investigated subtype-specific differences in Gag-protease-driven replication capacity using large well-characterized cohorts in Africa and the Americas. Patient-derived Gag-protease sequences were inserted into an HIV-1 NL4-3 backbone, and the replication capacities of the resulting recombinant viruses were measured in an HIV-1-inducible reporter T cell line by flow cytometry. Recombinant viruses expressing subtype C Gag-proteases exhibited substantially lower replication capacities than those expressing subtype B Gag-proteases (P < 0.0001); this observation remained consistent when representative Gag-protease sequences were engineered into an HIV-1 subtype C backbone. We identified Gag residues 483 and 484, located within the Alix-binding motif involved in virus budding, as major contributors to subtype-specific replicative differences. In East African cohorts, we observed a hierarchy of Gag-protease-driven replication capacities, i.e., subtypes A/C < D < intersubtype recombinants (P < 0.0029), which is consistent with reported intersubtype differences in disease progression. We thus hypothesize that the lower Gag-protease-driven replication capacity of subtypes A and C slows disease progression in individuals infected with these subtypes, which in turn leads to greater opportunity for transmission and thus increased prevalence of these subtypes.IMPORTANCE HIV-1 subtypes are unevenly distributed globally, and there are reported differences in their rates of disease progression and epidemic spread. The biological determinants underlying these differences have not been fully elucidated. Here, we show that HIV-1 Gag-protease-driven replication capacity correlates with the replication capacity of whole virus isolates. We further show that subtype B displays a significantly higher Gag-protease-mediated replication capacity than does subtype C, and we identify a major genetic determinant of these differences. Moreover, in two independent East African cohorts we demonstrate a reproducible hierarchy of Gag-protease-driven replicative capacity, whereby recombinants exhibit the greatest replication, followed by subtype D, followed by subtypes A and C. Our data identify Gag-protease as a major determinant of subtype differences in disease progression among HIV-1 subtypes; furthermore, we propose that the poorer viral replicative capacity of subtypes A and C may paradoxically contribute to their more efficient spread in sub-Saharan Africa.
Keywords: Gag-protease; HIV-1 subtype; viral replication capacity.
Copyright © 2017 American Society for Microbiology.
Figures


References
-
- Los Alamos National Laboratory. HIV sequence database. https://www.hiv.lanl.gov/content/sequence/HIV/mainpage.html Accessed 7 April 2014.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

