Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl
- PMID: 28424523
- PMCID: PMC6080695
- DOI: 10.1038/nature22063
Cohesin is positioned in mammalian genomes by transcription, CTCF and Wapl
Abstract
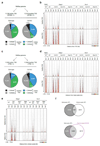
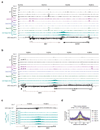
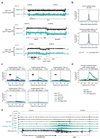
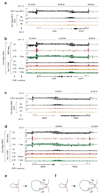
Mammalian genomes are spatially organized by CCCTC-binding factor (CTCF) and cohesin into chromatin loops and topologically associated domains, which have important roles in gene regulation and recombination. By binding to specific sequences, CTCF defines contact points for cohesin-mediated long-range chromosomal cis-interactions. Cohesin is also present at these sites, but has been proposed to be loaded onto DNA elsewhere and to extrude chromatin loops until it encounters CTCF bound to DNA. How cohesin is recruited to CTCF sites, according to this or other models, is unknown. Here we show that the distribution of cohesin in the mouse genome depends on transcription, CTCF and the cohesin release factor Wings apart-like (Wapl). In CTCF-depleted fibroblasts, cohesin cannot be properly recruited to CTCF sites but instead accumulates at transcription start sites of active genes, where the cohesin-loading complex is located. In the absence of both CTCF and Wapl, cohesin accumulates in up to 70 kilobase-long regions at 3'-ends of active genes, in particular if these converge on each other. Changing gene expression modulates the position of these 'cohesin islands'. These findings indicate that transcription can relocate mammalian cohesin over long distances on DNA, as previously reported for yeast cohesin, that this translocation contributes to positioning cohesin at CTCF sites, and that active genes can be freed from cohesin either by transcription-mediated translocation or by Wapl-mediated release.
Conflict of interest statement
The authors declare no competing financial interests.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

