Genetic basis for soma is present in undifferentiated volvocine green algae
- PMID: 28425150
- PMCID: PMC5540444
- DOI: 10.1111/jeb.13100
Genetic basis for soma is present in undifferentiated volvocine green algae
Abstract
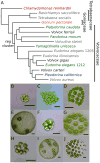
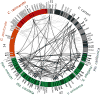
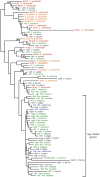
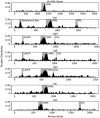
Somatic cellular differentiation plays a critical role in the transition from unicellular to multicellular life, but the evolution of its genetic basis remains poorly understood. By definition, somatic cells do not reproduce to pass on genes and so constitute an extreme form of altruistic behaviour. The volvocine green algae provide an excellent model system to study the evolution of multicellularity and somatic differentiation. In Volvox carteri, somatic cell differentiation is controlled by the regA gene, which is part of a tandem duplication of genes known as the reg cluster. Although previous work found the reg cluster in divergent Volvox species, its origin and distribution in the broader group of volvocine algae has not been known. Here, we show that the reg cluster is present in many species without somatic cells and determine that the genetic basis for soma arose before the phenotype at the origin of the family Volvocaceae approximately 200 million years ago. We hypothesize that the ancestral function was involved in regulating reproduction in response to stress and that this function was later co-opted to produce soma. Determining that the reg cluster was co-opted to control somatic cell development provides insight into how cellular differentiation, and with it greater levels of complexity and individuality, evolves.
Keywords: VARL domain; gene duplication; germ-soma differentiation; major transitions; multicellularity; volvocine algae.
© 2017 European Society For Evolutionary Biology. Journal of Evolutionary Biology © 2017 European Society For Evolutionary Biology.
Figures


Similar articles
-
Genome sequencing of the multicellular alga Astrephomene provides insights into convergent evolution of germ-soma differentiation.Sci Rep. 2021 Nov 22;11(1):22231. doi: 10.1038/s41598-021-01521-x. Sci Rep. 2021. PMID: 34811380 Free PMC article.
-
Characterization and Transformation of reg Cluster Genes in Volvox powersii Enable Investigation of Convergent Evolution of Cellular Differentiation in Volvox.Protist. 2021 Oct-Dec;172(5-6):125834. doi: 10.1016/j.protis.2021.125834. Epub 2021 Sep 20. Protist. 2021. PMID: 34695730
-
The Genetics of Fitness Reorganization during the Transition to Multicellularity: The Volvocine regA-like Family as a Model.Genes (Basel). 2023 Apr 19;14(4):941. doi: 10.3390/genes14040941. Genes (Basel). 2023. PMID: 37107699 Free PMC article. Review.
-
Early evolution of the genetic basis for soma in the volvocaceae.Evolution. 2014 Jul;68(7):2014-25. doi: 10.1111/evo.12416. Epub 2014 May 6. Evolution. 2014. PMID: 24689915
-
Evolution of reproductive development in the volvocine algae.Sex Plant Reprod. 2011 Jun;24(2):97-112. doi: 10.1007/s00497-010-0158-4. Epub 2010 Dec 21. Sex Plant Reprod. 2011. PMID: 21174128 Free PMC article. Review.
Cited by
-
Phylotranscriptomics points to multiple independent origins of multicellularity and cellular differentiation in the volvocine algae.BMC Biol. 2021 Aug 31;19(1):182. doi: 10.1186/s12915-021-01087-0. BMC Biol. 2021. PMID: 34465312 Free PMC article.
-
Evolution of Multicellular Complexity in The Dictyostelid Social Amoebas.Genes (Basel). 2021 Mar 27;12(4):487. doi: 10.3390/genes12040487. Genes (Basel). 2021. PMID: 33801615 Free PMC article. Review.
-
The Curious Case of Multicellularity in the Volvocine Algae.Front Genet. 2022 Feb 15;13:787665. doi: 10.3389/fgene.2022.787665. eCollection 2022. Front Genet. 2022. PMID: 35295942 Free PMC article. Review.
-
Genome sequencing of the multicellular alga Astrephomene provides insights into convergent evolution of germ-soma differentiation.Sci Rep. 2021 Nov 22;11(1):22231. doi: 10.1038/s41598-021-01521-x. Sci Rep. 2021. PMID: 34811380 Free PMC article.
-
Plasticity and the evolution of group-level regulation of cellular differentiation in the volvocine algae.Proc Biol Sci. 2025 Mar;292(2043):20242477. doi: 10.1098/rspb.2024.2477. Epub 2025 Mar 19. Proc Biol Sci. 2025. PMID: 40103550 Free PMC article.
References
-
- Akaike H. A New Look at the Statistical Model Identification. IEEE Trans Automat Contr. 1974;19:716–723.
-
- Bell G, Mooers AO. Size and complexity among multicellular organisms. Biol J Linn Soc. 1997;60:345–363.
-
- Burnham KP, Anderson DR. Model selection and multi-model inference: a practical information-theoretic approach. Springer-Verlag; New York: 2002.
-
- Buss L. The Evolution of Individuality. Princeton University Press; Princeton, NJ: 1987.
-
- Coleman AW. A Comparative Analysis of the Volvocaceae (Chlorophyta) J Phycol. 2012;48:491–513. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

