Hepatitis E Virus Genotypes and Evolution: Emergence of Camel Hepatitis E Variants
- PMID: 28425927
- PMCID: PMC5412450
- DOI: 10.3390/ijms18040869
Hepatitis E Virus Genotypes and Evolution: Emergence of Camel Hepatitis E Variants
Abstract
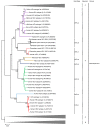
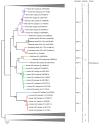
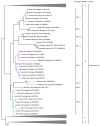
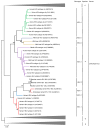
Hepatitis E virus (HEV) is a major cause of viral hepatitis globally. Zoonotic HEV is an important cause of chronic hepatitis in immunocompromised patients. The rapid identification of novel HEV variants and accumulating sequence information has prompted significant changes in taxonomy of the family Hepeviridae. This family includes two genera: Orthohepevirus, which infects terrestrial vertebrates, and Piscihepevirus, which infects fish. Within Orthohepevirus, there are four species, A-D, with widely differing host range. Orthohepevirus A contains the HEV variants infecting humans and its significance continues to expand with new clinical information. We now recognize eight genotypes within Orthohepevirus A: HEV1 and HEV2, restricted to humans; HEV3, which circulates among humans, swine, rabbits, deer and mongooses; HEV4, which circulates between humans and swine; HEV5 and HEV6, which are found in wild boars; and HEV7 and HEV8, which were recently identified in dromedary and Bactrian camels, respectively. HEV7 is an example of a novel genotype that was found to have significance to human health shortly after discovery. In this review, we summarize recent developments in HEV molecular taxonomy, epidemiology and evolution and describe the discovery of novel camel HEV genotypes as an illustrative example of the changes in this field.
Keywords: Bactrian camel; dromedary camel; genotypes; hepatitis; hepatitis E; molecular epidemiology; molecular evolution; swine; taxonomy; zoonosis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- WHO Hepatitis E Fact Sheet. [(accessed on 15 February 2017)]; Available online: http://www.who.int/mediacentre/factsheets/fs280/en/
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

