Seasonal Dynamics of Haptophytes and dsDNA Algal Viruses Suggest Complex Virus-Host Relationship
- PMID: 28425942
- PMCID: PMC5408690
- DOI: 10.3390/v9040084
Seasonal Dynamics of Haptophytes and dsDNA Algal Viruses Suggest Complex Virus-Host Relationship
Abstract
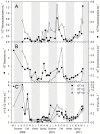
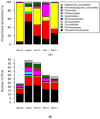
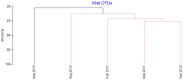
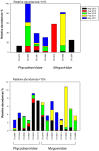
Viruses influence the ecology and diversity of phytoplankton in the ocean. Most studies of phytoplankton host-virus interactions have focused on bloom-forming species like Emiliania huxleyi or Phaeocystis spp. The role of viruses infecting phytoplankton that do not form conspicuous blooms have received less attention. Here we explore the dynamics of phytoplankton and algal viruses over several sequential seasons, with a focus on the ubiquitous and diverse phytoplankton division Haptophyta, and their double-stranded DNA viruses, potentially with the capacity to infect the haptophytes. Viral and phytoplankton abundance and diversity showed recurrent seasonal changes, mainly explained by hydrographic conditions. By 454 tag-sequencing we revealed 93 unique haptophyte operational taxonomic units (OTUs), with seasonal changes in abundance. Sixty-one unique viral OTUs, representing Megaviridae and Phycodnaviridae, showed only distant relationship with currently isolated algal viruses. Haptophyte and virus community composition and diversity varied substantially throughout the year, but in an uncoordinated manner. A minority of the viral OTUs were highly abundant at specific time-points, indicating a boom-bust relationship with their host. Most of the viral OTUs were very persistent, which may represent viruses that coexist with their hosts, or able to exploit several host species.
Keywords: Haptophyta; Megaviridae; Phycodnaviridae; marine viral ecology; metagenomics; viral–host interactions.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Eikrem E., Medlin L.K., Henderiks J., Rokitta S., Rost B., Probert I., Throndsen J., Edvardsen B. Haptophyta. In: Archibald J.M., Simpson A.G.B., Slamovits C.H., Margulis L., Melkonian M., Chapman D.J., Corliss J.O., editors. Handbook of the Protists. Springer International Publishing; Cham, Switzerland: 2016. pp. 1–61.
-
- Hallegraeff G.M. Ocean climate change, phytoplankton community responses, and harmful algal blooms: A formidable predictive challenge. J. Phycol. 2010;46:220–235. doi: 10.1111/j.1529-8817.2010.00815.x. - DOI
-
- Leadbeater B.S.C. Identification, by means of electron microscopy, of flagellate nanoplankton from the coast of Norway. Sarsia. 1972;49:107–124. doi: 10.1080/00364827.1972.10411212. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

