Effects of over-expressing a native gene encoding 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) on glyphosate resistance in Arabidopsis thaliana
- PMID: 28426703
- PMCID: PMC5398549
- DOI: 10.1371/journal.pone.0175820
Effects of over-expressing a native gene encoding 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) on glyphosate resistance in Arabidopsis thaliana
Abstract
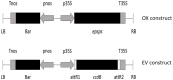
Widespread overuse of the herbicide glyphosate, the active ingredient in RoundUp®, has led to the evolution of glyphosate-resistant weed biotypes, some of which persist by overproducing the herbicide's target enzyme, 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS). EPSPS is a key enzyme in the shikimic acid pathway for biosynthesis of aromatic amino acids, lignin, and defensive compounds, but little is known about how overproducing EPSPS affects downstream metabolites, growth, or lifetime fitness in the absence of glyphosate. We are using Arabidopsis as a model system for investigating phenotypic effects of overproducing EPSPS, thereby avoiding confounding effects of genetic background or other mechanisms of herbicide resistance in agricultural weeds. Here, we report results from the first stage of this project. We designed a binary vector expressing a native EPSPS gene from Arabidopsis under control of the CaMV35S promoter (labelled OX, for over-expression). For both OX and the empty vector (labelled EV), we obtained nine independent T3 lines. Subsets of these lines were used to characterize glyphosate resistance in greenhouse experiments. Seven of the nine OX lines exhibited enhanced glyphosate resistance when compared to EV and wild-type control lines, and one of these was discarded due to severe deformities. The remaining six OX lines exhibited enhanced EPSPS gene expression and glyphosate resistance compared to controls. Glyphosate resistance was correlated with the degree of EPSPS over-expression for both vegetative and flowering plants, indicating that glyphosate resistance can be used as a surrogate for EPSPS expression levels in this system. These findings set the stage for examination of the effects of EPSPS over-expression on fitness-related traits in the absence of glyphosate. We invite other investigators to contact us if they wish to study gene expression, downstream metabolic effects, and other questions with these particular lines.
Conflict of interest statement
Figures



Similar articles
-
An intragenic approach to confer glyphosate resistance in chile (Capsicum annuum) by introducing an in vitro mutagenized chile EPSPS gene encoding for a glyphosate resistant EPSPS protein.PLoS One. 2018 Apr 12;13(4):e0194666. doi: 10.1371/journal.pone.0194666. eCollection 2018. PLoS One. 2018. PMID: 29649228 Free PMC article.
-
Evaluation of glyphosate resistance in Arabidopsis thaliana expressing an altered target site EPSPS.Pest Manag Sci. 2018 May;74(5):1174-1183. doi: 10.1002/ps.4654. Epub 2017 Sep 7. Pest Manag Sci. 2018. PMID: 28677849 Free PMC article.
-
The Triple Amino Acid Substitution TAP-IVS in the EPSPS Gene Confers High Glyphosate Resistance to the Superweed Amaranthus hybridus.Int J Mol Sci. 2019 May 15;20(10):2396. doi: 10.3390/ijms20102396. Int J Mol Sci. 2019. PMID: 31096560 Free PMC article.
-
Glyphosate Resistance and EPSPS Gene Duplication: Convergent Evolution in Multiple Plant Species.J Hered. 2018 Feb 14;109(2):117-125. doi: 10.1093/jhered/esx087. J Hered. 2018. PMID: 29040588 Review.
-
Deciphering the Mechanism of Glyphosate Resistance in Amaranthus palmeri by Cytogenomics.Cytogenet Genome Res. 2021;161(12):578-584. doi: 10.1159/000521409. Epub 2022 Jan 12. Cytogenet Genome Res. 2021. PMID: 35021177 Review.
Cited by
-
An intragenic approach to confer glyphosate resistance in chile (Capsicum annuum) by introducing an in vitro mutagenized chile EPSPS gene encoding for a glyphosate resistant EPSPS protein.PLoS One. 2018 Apr 12;13(4):e0194666. doi: 10.1371/journal.pone.0194666. eCollection 2018. PLoS One. 2018. PMID: 29649228 Free PMC article.
-
Overexpressing Exogenous 5-Enolpyruvylshikimate-3-Phosphate Synthase (EPSPS) Genes Increases Fecundity and Auxin Content of Transgenic Arabidopsis Plants.Front Plant Sci. 2018 Feb 27;9:233. doi: 10.3389/fpls.2018.00233. eCollection 2018. Front Plant Sci. 2018. PMID: 29535747 Free PMC article.
-
Root Microbiome Modulates Plant Growth Promotion Induced by Low Doses of Glyphosate.mSphere. 2020 Aug 12;5(4):e00484-20. doi: 10.1128/mSphere.00484-20. mSphere. 2020. PMID: 32817451 Free PMC article.
-
Increased Longevity and Dormancy of Soil-Buried Seeds from Advanced Crop-Wild Rice Hybrids Overexpressing the EPSPS Transgene.Biology (Basel). 2021 Jun 20;10(6):562. doi: 10.3390/biology10060562. Biology (Basel). 2021. PMID: 34203092 Free PMC article.
-
Reduced Carbon Dioxide by Overexpressing EPSPS Transgene in Arabidopsis and Rice: Implications in Carbon Neutrality through Genetically Engineered Plants.Biology (Basel). 2023 Dec 31;13(1):25. doi: 10.3390/biology13010025. Biology (Basel). 2023. PMID: 38248456 Free PMC article.
References
-
- Franz JE, Mao MK, Sikorski JA. Glyphosate: a unique global herbicide. Washington, DC, USA: American Chemistry Society; 1997.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

