The influence of cell membrane and SNAP25 linker loop on the dynamics and unzipping of SNARE complex
- PMID: 28426820
- PMCID: PMC5398687
- DOI: 10.1371/journal.pone.0176235
The influence of cell membrane and SNAP25 linker loop on the dynamics and unzipping of SNARE complex
Abstract
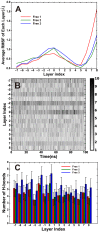
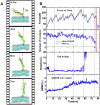
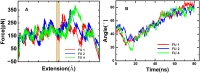
The soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) complex is composed of three neuronal proteins VAMP2, Syntaxin and SNAP25, which plays a core role during the process of membrane fusion. The zipping assembly of the SNARE complex releases energies and drives the vesicle and cell membrane into close proximity. In this study, we use all-atom molecular dynamics simulations to probe the dynamics of SNARE and its unzipping process in the context of membrane at the atomistic details. Our results indicated that the NTD of SNARE core domain is relatively more stable than CTD, which is in agreement with previous experiments. More importantly, possible interactions between the linker loop (LL) region of SNAP25 and VAMP2 are observed, suggests that the LL region may facilitate VAMP2 binding and SNARE initiation. The forced unzipping of SNARE in the presence of membrane and LL of SNAP25 reveals the possible pathway for energy generation of SNARE zipping, provides information to understand how force may regulate the cooperativity between the membrane and the SNARE complex.
Conflict of interest statement
Figures



Similar articles
-
Membrane-mediated disorder-to-order transition of SNAP25 flexible linker facilitates its interaction with syntaxin-1 and SNARE-complex assembly.FASEB J. 2019 Jul;33(7):7985-7994. doi: 10.1096/fj.201802796R. Epub 2019 Mar 27. FASEB J. 2019. PMID: 30916996
-
All-atom and coarse-grained simulations of the forced unfolding pathways of the SNARE complex.Proteins. 2014 Jul;82(7):1376-86. doi: 10.1002/prot.24505. Epub 2014 Feb 6. Proteins. 2014. PMID: 24403006
-
Functionally distinct SNARE motifs of SNAP25 cooperate in SNARE assembly and membrane fusion.Biophys J. 2025 Feb 18;124(4):637-650. doi: 10.1016/j.bpj.2024.12.034. Epub 2024 Dec 31. Biophys J. 2025. PMID: 39982442 Free PMC article.
-
Neuronal SNARE complex: A protein folding system with intricate protein-protein interactions, and its common neuropathological hallmark, SNAP25.Neurochem Int. 2019 Jan;122:196-207. doi: 10.1016/j.neuint.2018.12.001. Epub 2018 Dec 2. Neurochem Int. 2019. PMID: 30517887 Review.
-
A role for V-ATPase subunits in synaptic vesicle fusion?J Neurochem. 2011 May;117(4):603-12. doi: 10.1111/j.1471-4159.2011.07234.x. Epub 2011 Mar 28. J Neurochem. 2011. PMID: 21375531 Review.
Cited by
-
Long-Term Safety and Clinical Effects of Nilotinib in Parkinson's Disease.Mov Disord. 2021 Mar;36(3):740-749. doi: 10.1002/mds.28389. Epub 2020 Nov 20. Mov Disord. 2021. PMID: 33215762 Free PMC article. Clinical Trial.
-
MicroRNA-210-5p Contributes to Cognitive Impairment in Early Vascular Dementia Rat Model Through Targeting Snap25.Front Mol Neurosci. 2018 Nov 13;11:388. doi: 10.3389/fnmol.2018.00388. eCollection 2018. Front Mol Neurosci. 2018. PMID: 30483048 Free PMC article.
-
Discoidin Domain Receptor 1 is a therapeutic target for neurodegenerative diseases.Hum Mol Genet. 2020 Oct 10;29(17):2882-2898. doi: 10.1093/hmg/ddaa177. Hum Mol Genet. 2020. PMID: 32776088 Free PMC article.
-
The SNAP-25 linker supports fusion intermediates by local lipid interactions.Elife. 2019 Mar 18;8:e41720. doi: 10.7554/eLife.41720. Elife. 2019. PMID: 30883328 Free PMC article.
-
Multiple structural states in an intrinsically disordered protein, SNAP-25, using circular dichroism.Biophys J. 2025 Jun 3;124(11):1828-1842. doi: 10.1016/j.bpj.2025.02.003. Epub 2025 Feb 8. Biophys J. 2025. PMID: 39923128 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

